5IAG
 
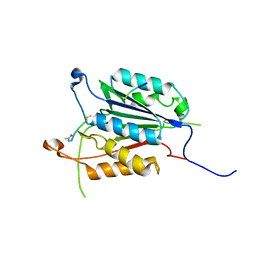 | | Caspase 3 V266Q | | Descriptor: | ACE-ASP-GLU-VAL-ASK, ASP-ASP-ASP-MET, Caspase-3 | | Authors: | Maciag, J.J, Mackenzie, S.H, Tucker, M.B, Schipper, J.L, Swartz, P.D, Clark, A.C. | | Deposit date: | 2016-02-21 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Tunable allosteric library of caspase-3 identifies coupling between conserved water molecules and conformational selection.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5I8Q
 
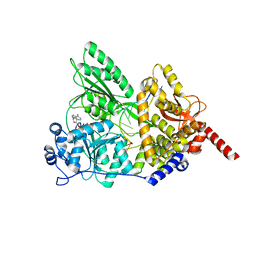 | | S. cerevisiae Prp43 in complex with RNA and ADPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP43, ... | | Authors: | He, Y, Nielsen, K.H, Andersen, G.R. | | Deposit date: | 2016-02-19 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structure of the DEAH/RHA ATPase Prp43p bound to RNA implicates a pair of hairpins and motif Va in translocation along RNA.
RNA, 23, 2017
|
|
5IDR
 
 | |
2NCV
 
 | | NMR structure of RWS21 structure in LPS micelles | | Descriptor: | Heparin cofactor 2 | | Authors: | Datta, A, Bhunia, A. | | Deposit date: | 2016-04-18 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Effects of N-Terminal Modifications of the Anti-Inflammatory Peptide KYE21 on Lipopolysaccharide and Membrane Interactions
To be Published
|
|
5IEW
 
 | |
6OQC
 
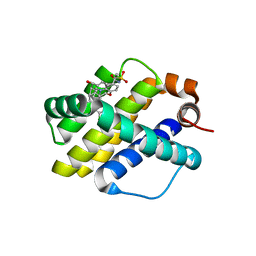 | | Crystal structure of Mcl1 with inhibitor 9 | | Descriptor: | (4S,7aR,9aR,10S,11E,15R)-6'-chloro-10-hydroxy-15-methyl-3',4',7a,8,9,9a,10,13,14,15-decahydro-2'H,3H,5H-spiro[1,19-(ethanediylidene)-16lambda~6~-cyclobuta[i][1,4]oxazepino[3,4-f][1,2,7]thiadiazacyclohexadecine-4,1'-naphthalene]-16,16,18(7H,17H)-trione, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X. | | Deposit date: | 2019-04-26 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | AMG 176, a Selective MCL1 Inhibitor, Is Effective in Hematologic Cancer Models Alone and in Combination with Established Therapies.
CANCER DISCOV, 8, 2018
|
|
2NN6
 
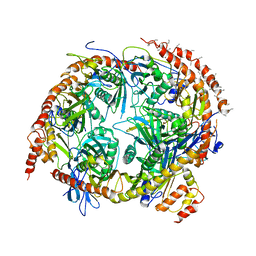 | | Structure of the human RNA exosome composed of Rrp41, Rrp45, Rrp46, Rrp43, Mtr3, Rrp42, Csl4, Rrp4, and Rrp40 | | Descriptor: | 3'-5' exoribonuclease CSL4 homolog, Exosome complex exonuclease RRP4, Exosome complex exonuclease RRP40, ... | | Authors: | Lima, C.D. | | Deposit date: | 2006-10-23 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Reconstitution, activities, and structure of the eukaryotic RNA exosome.
Cell(Cambridge,Mass.), 127, 2006
|
|
5MFW
 
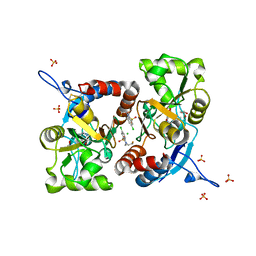 | | Crystal structure of the GluK1 ligand-binding domain in complex with kainate and BPAM-121 at 2.10 A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, 7-chloro-4-(2-fluoroethyl)-2,3-dihydro-1,2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, ... | | Authors: | Larsen, A.P, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2016-11-18 | | Release date: | 2017-04-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and Structure-Function Study of Positive Allosteric Modulators of Kainate Receptors.
Mol. Pharmacol., 91, 2017
|
|
6A5J
 
 | | solution NMR Structure of small peptide | | Descriptor: | ILE-LYS-LYS-ILE-LEU-SER-LYS-ILE-LYS-LYS-LEU-LEU-LYS | | Authors: | Feng, L.B, Dong, W.B. | | Deposit date: | 2018-06-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution NMR Structure of antimicrobial peptide L-K6, the analog of temporin-1CEB derived from the skin secretions of Chinese brown frog Rana chensinenesis
To Be Published
|
|
6ALK
 
 | | NMR solution structure of the major beech pollen allergen Fag s 1 | | Descriptor: | Fag s 1 pollen allergen | | Authors: | Moraes, A.H, Asam, A, Almeida, F.C.L, Wallner, M, Ferreira, F, Valente, A.P. | | Deposit date: | 2017-08-08 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for cross-reactivity and conformation fluctuation of the major beech pollen allergen Fag s 1.
Sci Rep, 8, 2018
|
|
1ZS5
 
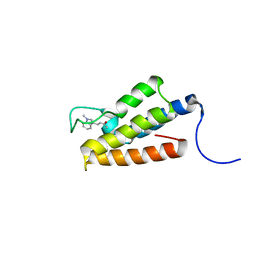 | | Structure-based evaluation of selective and non-selective small molecules that block HIV-1 TAT and PCAF association | | Descriptor: | (3E)-4-(1-METHYL-1H-INDOL-3-YL)BUT-3-EN-2-ONE, Histone acetyltransferase PCAF | | Authors: | Zeng, L, Godbole, S, Muller, M, Yan, S, Sanchez, R, Zhou, M. | | Deposit date: | 2005-05-23 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure-based evaluation of selective nad non-selective small molecules that block hiv-1 tat and pcaf association
TO BE PUBLISHED
|
|
6AFQ
 
 | |
5ION
 
 | |
5MMU
 
 | | NMR solution structure of the major apple allergen Mal d 1 | | Descriptor: | Major allergen Mal d 1 | | Authors: | Ahammer, L, Grutsch, S, Kamenik, A.S, Liedl, K.R, Tollinger, M. | | Deposit date: | 2016-12-12 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Major Apple Allergen Mal d 1.
J. Agric. Food Chem., 65, 2017
|
|
5IEC
 
 | |
7UQ3
 
 | | JmjC domain-containing protein 5 (JMJD5) in complex with Mn and (S)-2-(1-hydroxy-2,5-dioxopyrrolidin-3-yl)acetic acid | | Descriptor: | Bifunctional peptidase and arginyl-hydroxylase JMJD5, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Chowdhury, R, Islam, M.S, Schofield, C.J. | | Deposit date: | 2022-04-19 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural analysis of the 2-oxoglutarate binding site of the circadian rhythm linked oxygenase JMJD5.
Sci Rep, 12, 2022
|
|
6ACV
 
 | | the solution NMR structure of MBD domain | | Descriptor: | Methyl-CpG-binding domain-containing protein 11 | | Authors: | Li, S.L, Feng, Y.Y, Zhou, Y, Ding, Y.M, Liu, K, Nie, Y, Li, F, Yang, Y.Y. | | Deposit date: | 2018-07-27 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | the solution NMR structure of MBD domains
To Be Published
|
|
6AAS
 
 | | Solution Structure for helix 45 in 3' end of 12S rRNA | | Descriptor: | RNA (28-MER) | | Authors: | Liu, X, Wu, P. | | Deposit date: | 2018-07-19 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
3EX3
 
 | | human orotidyl-5'-monophosphate decarboxylase in complex with 6-azido-UMP, covalent adduct | | Descriptor: | GLYCEROL, Orotidine-5'-phosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Heinrich, D, Diederichsen, U, Rudolph, M. | | Deposit date: | 2008-10-16 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Lys314 is a nucleophile in non-classical reactions of orotidine-5'-monophosphate decarboxylase
Chemistry, 15, 2009
|
|
5J4Z
 
 | | Architecture of tight respirasome | | Descriptor: | COMPLEX I 13KDA/NDUFS6, COMPLEX I 15KDA/NDUFS5, COMPLEX I 18KDA/NDUFS6, ... | | Authors: | Letts, J.A, Fiedorczuk, K, Sazanov, L.A. | | Deposit date: | 2016-04-01 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | The architecture of respiratory supercomplexes.
Nature, 537, 2016
|
|
6AQU
 
 | | Crystal Structure of Plasmodium falciparum purine nucleoside phosphorylase: The M183L mutant | | Descriptor: | PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Harijan, R.K, Ducati, R.G, Namanja-Magliano, H.A, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-08-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Genetic resistance to purine nucleoside phosphorylase inhibition inPlasmodium falciparum.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5J7C
 
 | |
3JYI
 
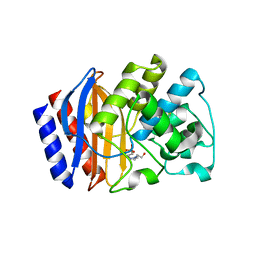 | | Structural and biochemical evidence that a TEM-1 {beta}-lactamase Asn170Gly active site mutant acts via substrate-assisted catalysis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase TEM, PHOSPHATE ION | | Authors: | Brown, N.G, Palzkill, T.G, Prasad, B.V.V, Shanker, S. | | Deposit date: | 2009-09-21 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structural and biochemical evidence that a TEM-1 beta-lactamase N170G active site mutant acts via substrate-assisted catalysis
J.Biol.Chem., 284, 2009
|
|
2N8J
 
 | |
6AUV
 
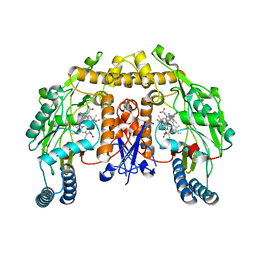 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 4-Methyl-6-(2-(5-(3-((methylamino)methyl)phenyl)pyridin-3-yl)ethyl)pyridin-2-amine | | Descriptor: | 4-methyl-6-[2-(5-{3-[(methylamino)methyl]phenyl}pyridin-3-yl)ethyl]pyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Improvement of Cell Permeability of Human Neuronal Nitric Oxide Synthase Inhibitors Using Potent and Selective 2-Aminopyridine-Based Scaffolds with a Fluorobenzene Linker.
J. Med. Chem., 60, 2017
|
|
