3HM3
 
 | |
5ZC4
 
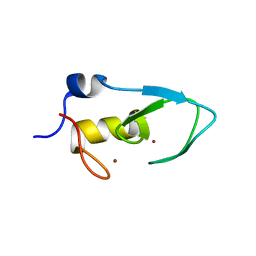 | |
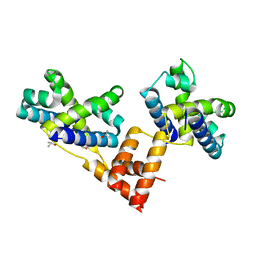
5ZBU
 
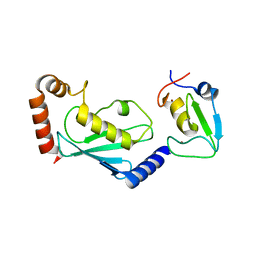 | |
1XWR
 
 | | Crystal structure of the coliphage lambda transcription activator protein CII | | Descriptor: | ISOPROPYL ALCOHOL, Regulatory protein CII | | Authors: | Datta, A.B, Panjikar, S, Weiss, M.S, Chakrabarti, P, Parrack, P. | | Deposit date: | 2004-11-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure of {lambda} CII: Implications for recognition of direct-repeat DNA by an unusual tetrameric organization
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
5WRX
 
 | | VG13P structure in LPS | | Descriptor: | analogue peptide VG13P | | Authors: | Datta, A, Bhunia, A. | | Deposit date: | 2016-12-04 | | Release date: | 2017-05-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and Dynamic Insights into a Glycine-Mediated Short Analogue of a Designed Peptide in Lipopolysaccharide Micelles: Correlation Between Compact Structure and Anti-Endotoxin Activity.
Biochemistry, 56, 2017
|
|
5XRX
 
 | | EFK17DA structure in Microgel MAA60 | | Descriptor: | Cathelicidin antimicrobial peptide | | Authors: | Datta, A, Bhunia, A. | | Deposit date: | 2017-06-10 | | Release date: | 2018-04-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformational Aspects of High Content Packing of Antimicrobial Peptides in Polymer Microgels
ACS Appl Mater Interfaces, 9, 2017
|
|
6M0U
 
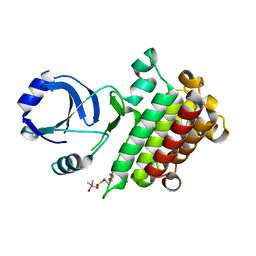 | |
5XNG
 
 | | EFK17A structure in Microgel MAA60 | | Descriptor: | Cathelicidin antimicrobial peptide | | Authors: | Datta, A, Bhunia, A. | | Deposit date: | 2017-05-22 | | Release date: | 2018-04-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformational Aspects of High Content Packing of Antimicrobial Peptides in Polymer Microgels
ACS Appl Mater Interfaces, 9, 2017
|
|
2NCW
 
 | |
2NCU
 
 | |
2N65
 
 | | Disulphide linked homodimer of designed antimicrobial peptide VG16KRKP | | Descriptor: | antimicrobial peptide VG16KRKP | | Authors: | Datta, A, Bhunia, A. | | Deposit date: | 2015-08-12 | | Release date: | 2016-03-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Designing potent antimicrobial peptides by disulphide linked dimerization and N-terminal lipidation to increase antimicrobial activity and membrane perturbation: Structural insights into lipopolysaccharide binding.
J Colloid Interface Sci, 461, 2016
|
|
2NCV
 
 | | NMR structure of RWS21 structure in LPS micelles | | Descriptor: | Heparin cofactor 2 | | Authors: | Datta, A, Bhunia, A. | | Deposit date: | 2016-04-18 | | Release date: | 2017-04-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Effects of N-Terminal Modifications of the Anti-Inflammatory Peptide KYE21 on Lipopolysaccharide and Membrane Interactions
To be Published
|
|
3BT8
 
 | |
4S1E
 
 | |
4S1J
 
 | |
3EOV
 
 | | Crystal structure of cyclophilin from Leishmania donovani ligated with cyclosporin A | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Venugopal, V, Dasgupta, D, Datta, A.K, Banerjee, R. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Cyclophilin from Leishmania Donovani Bound to Cyclosporin at 2.6 A Resolution: Correlation between Structure and Thermodynamic Data.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4K2E
 
 | | HlyU from Vibrio cholerae N16961 | | Descriptor: | Transcriptional activator HlyU | | Authors: | Mukherjee, D, Datta, A.B, Chakrabarti, P. | | Deposit date: | 2013-04-09 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of HlyU, the hemolysin gene transcription activator, from Vibrio cholerae N16961 and functional implications.
Biochim.Biophys.Acta, 1844, 2014
|
|
2HAQ
 
 | | Crystal Structure of Cyclophilin A from Leishmania Donovani | | Descriptor: | Cyclophilin | | Authors: | Venugopal, V, Sen, B, Datta, A.K, Banerjee, R. | | Deposit date: | 2006-06-13 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of cyclophilin from Leishmania donovani at 1.97 A resolution.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
6L2B
 
 | |
5YWR
 
 | | Crystal Structure of RING E3 ligase ZNRF1 in complex with Ube2N (Ubc13) | | Descriptor: | E3 ubiquitin-protein ligase ZNRF1, FORMIC ACID, TRIETHYLENE GLYCOL, ... | | Authors: | Behera, A.P, Naskar, P, Datta, A.B. | | Deposit date: | 2017-11-30 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural insights into the nanomolar affinity of RING E3 ligase ZNRF1 for Ube2N and its functional implications.
Biochem. J., 475, 2018
|
|
3MHS
 
 | | Structure of the SAGA Ubp8/Sgf11/Sus1/Sgf73 DUB module bound to ubiquitin aldehyde | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein SUS1, ... | | Authors: | Samara, N.L, Datta, A.B, Berndsen, C.E, Zhang, X, Yao, T, Cohen, R.E, Wolberger, C. | | Deposit date: | 2010-04-08 | | Release date: | 2010-04-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural insights into the assembly and function of the SAGA deubiquitinating module.
Science, 328, 2010
|
|
3MHH
 
 | | Structure of the SAGA Ubp8/Sgf11/Sus1/Sgf73 DUB module | | Descriptor: | Protein SUS1, SAGA-associated factor 11, SAGA-associated factor 73, ... | | Authors: | Samara, N.L, Datta, A.B, Berndsen, C.E, Zhang, X, Yao, T, Cohen, R.E, Wolberger, C. | | Deposit date: | 2010-04-08 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights into the assembly and function of the SAGA deubiquitinating module.
Science, 328, 2010
|
|
2N63
 
 | | Structure of C4VG16KRKP | | Descriptor: | PENTANOIC ACID, antimicrobial peptide C4VG16KRKP | | Authors: | Bhunia, A, Datta, A. | | Deposit date: | 2015-08-11 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-24 | | Method: | SOLUTION NMR | | Cite: | Designing potent antimicrobial peptides by disulphide linked dimerization and N-terminal lipidation to increase antimicrobial activity and membrane perturbation: Structural insights into lipopolysaccharide binding.
J Colloid Interface Sci, 461, 2016
|
|
4OOI
 
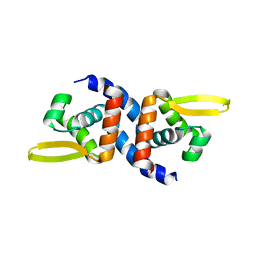 | | Reduced HlyU from Vibrio cholerae N16961 | | Descriptor: | Transcriptional activator HlyU | | Authors: | Mukherjee, D, Datta, A.B, Chakrabarti, P. | | Deposit date: | 2014-02-03 | | Release date: | 2014-12-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of HlyU, the hemolysin gene transcription activator, from Vibrio cholerae N16961 and functional implications
Biochim.Biophys.Acta, 1844, 2014
|
|
2MWL
 
 | |
