6MMU
 
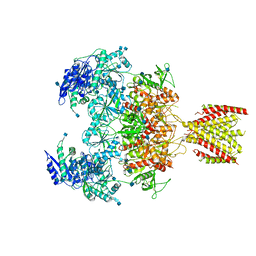 | | Triheteromeric NMDA receptor GluN1/GluN2A/GluN2A* in the '2-Knuckle-Asymmetric' conformation, in complex with glycine and glutamate, in the presence of 1 micromolar zinc chloride, and at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Jalali-Yazdi, F, Chowdhury, S, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-10-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor.
Cell, 175, 2018
|
|
5HZK
 
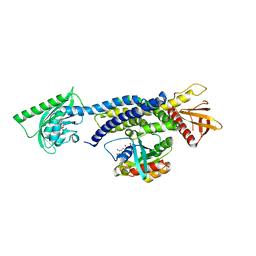 | | Crystal structure of photoinhibitable Intersectin1 containing wildtype LOV2 domain in complex with Cdc42 | | Descriptor: | Cell division control protein 42 homolog, FLAVIN MONONUCLEOTIDE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tarnawski, M, Dagliyan, O, Chu, P.H, Shirvanyants, D, Dokholyan, N.V, Hahn, K.M, Schlichting, I. | | Deposit date: | 2016-02-02 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Engineering extrinsic disorder to control protein activity in living cells.
Science, 354, 2016
|
|
6V0O
 
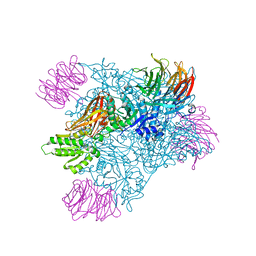 | | PRMT5 bound to the PBM peptide from pICln | | Descriptor: | ACETYL GROUP, Methylosome protein 50, PBM peptide, ... | | Authors: | McMillan, B.J, Raymond, D.D. | | Deposit date: | 2019-11-19 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Molecular basis for substrate recruitment to the PRMT5 methylosome.
Mol.Cell, 81, 2021
|
|
8ESK
 
 | | Cryo-EM structure of Torpedo nicotinic acetylcholine receptor in complex with rocuronium, resting-like state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goswami, U, Rahman, M.M, Teng, J, Hibbs, R.E. | | Deposit date: | 2022-10-14 | | Release date: | 2023-06-07 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural interplay of anesthetics and paralytics on muscle nicotinic receptors.
Nat Commun, 14, 2023
|
|
3LGT
 
 | |
6N23
 
 | |
6N28
 
 | | BEST1 calcium-bound open state | | Descriptor: | Bestrophin homolog, CALCIUM ION | | Authors: | Miller, A.N, Vaisey, G, Long, S.B. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanisms of gating in the calcium-activated chloride channel bestrophin.
Elife, 8, 2019
|
|
3IOL
 
 | |
6NBB
 
 | |
3ATJ
 
 | | HEME LIGAND MUTANT OF RECOMBINANT HORSERADISH PEROXIDASE IN COMPLEX WITH BENZHYDROXAMIC ACID | | Descriptor: | BENZHYDROXAMIC ACID, CALCIUM ION, PROTEIN (HORSERADISH PEROXIDASE C1A), ... | | Authors: | Meno, K, White, C.G, Smith, A.T, Gajhede, M. | | Deposit date: | 1998-12-16 | | Release date: | 1999-04-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Catalytical Implications of a F221M Mutation in the Proximal Pocket of Horseradish Peroxidase C (HRP C)
To be Published
|
|
3ILY
 
 | |
3AVQ
 
 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with N9-Pan | | Descriptor: | (2S)-2,4-dihydroxy-3,3-dimethyl-N-[3-(nonylamino)-3-oxopropyl]butanamide, CITRATE ANION, GLYCEROL, ... | | Authors: | Chetnani, B, Kumar, P, Abhinav, K.V, Chhibber, M, Surolia, A, Vijayan, M. | | Deposit date: | 2011-03-06 | | Release date: | 2011-08-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Location and conformation of pantothenate and its derivatives in Mycobacterium tuberculosis pantothenate kinase: insights into enzyme action
Acta Crystallogr.,Sect.D, 67, 2011
|
|
5IPT
 
 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 5 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (14.1 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
6CPP
 
 | | CRYSTAL STRUCTURES OF CYTOCHROME P450-CAM COMPLEXED WITH CAMPHANE, THIOCAMPHOR, AND ADAMANTANE: FACTORS CONTROLLING P450 SUBSTRATE HYDROXYLATION | | Descriptor: | CAMPHANE, CYTOCHROME P450-CAM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Raag, R, Poulos, T.L. | | Deposit date: | 1990-05-18 | | Release date: | 1991-07-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of cytochrome P-450CAM complexed with camphane, thiocamphor, and adamantane: factors controlling P-450 substrate hydroxylation.
Biochemistry, 30, 1991
|
|
3GEN
 
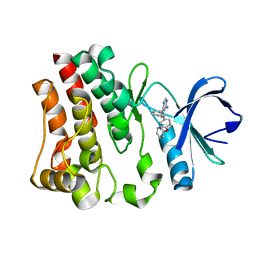 | | The 1.6 A crystal structure of human bruton's tyrosine kinase bound to a pyrrolopyrimidine-containing compound | | Descriptor: | 4-Amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl-cyclopentane, Tyrosine-protein kinase BTK | | Authors: | Marcotte, D.J, Liu, Y.T, Arduini, R.M, Hession, C.A, Miatkowski, K, Wildes, C.P, Cullen, P.F, Hopkins, B, Mertsching, E, Jenkins, T.J, Romanowski, M.J, Baker, D.P, Silvian, L.F. | | Deposit date: | 2009-02-25 | | Release date: | 2010-01-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of human Bruton's tyrosine kinase in active and inactive conformations suggest a mechanism of activation for TEC family kinases.
Protein Sci., 19, 2010
|
|
8R4U
 
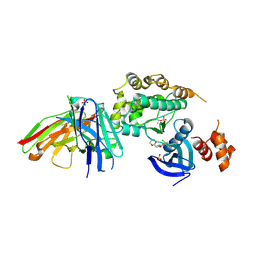 | | Structure of salt-inducible kinase 3 with inhibitors | | Descriptor: | 8-[(5-azanyl-1,3-dioxan-2-yl)methyl]-6-[2-chloranyl-4-(3-fluoranylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, SULFATE ION, Serine/threonine-protein kinase SIK3, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2023-11-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.416 Å) | | Cite: | The structures of salt-inducible kinase 3 in complex with inhibitors reveal determinants for binding and selectivity.
J.Biol.Chem., 300, 2024
|
|
7L2J
 
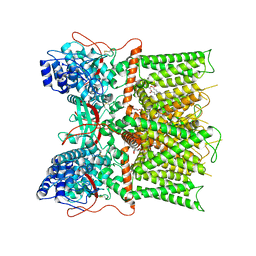 | | Cryo-EM structure of full-length TRPV1 at pH6c state | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7L2I
 
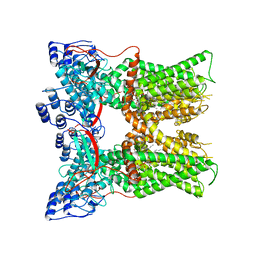 | | Cryo-EM structure of full-length TRPV1 at pH6a state | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1S,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7L2K
 
 | |
1RD7
 
 | | DIHYDROFOLATE REDUCTASE COMPLEXED WITH FOLATE | | Descriptor: | BETA-MERCAPTOETHANOL, DIHYDROFOLATE REDUCTASE, FOLIC ACID | | Authors: | Sawaya, M.R, Kraut, J. | | Deposit date: | 1996-11-01 | | Release date: | 1996-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: crystallographic evidence.
Biochemistry, 36, 1997
|
|
7L2M
 
 | | Cryo-EM structure of DkTx/RTX-bound full-length TRPV1 | | Descriptor: | SODIUM ION, Tau-theraphotoxin-Hs1a, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
4FF6
 
 | | Mycobacterium tuberculosis DprE1 in complex with CT325 - monoclinic crystal form | | Descriptor: | 3-(hydroxyamino)-N-[(1R)-1-phenylethyl]-5-(trifluoromethyl)benzamide, FLAVIN-ADENINE DINUCLEOTIDE, IMIDAZOLE, ... | | Authors: | Batt, S.M, Besra, G.S, Futterer, K. | | Deposit date: | 2012-05-31 | | Release date: | 2012-07-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of inhibition of Mycobacterium tuberculosis DprE1 by benzothiazinone inhibitors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8R4Q
 
 | | Salt inducible kinase 3 in complex with inhibitor | | Descriptor: | 4-[(2,4-dichloro-5-methoxyphenyl)amino]-6-methoxy-7-[3-(4-methylpiperazin-1-yl)propoxy]quinoline-3-carbonitrile, SULFATE ION, Serine/threonine-protein kinase SIK3, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2023-11-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.838 Å) | | Cite: | The structures of salt-inducible kinase 3 in complex with inhibitors reveal determinants for binding and selectivity.
J.Biol.Chem., 300, 2024
|
|
8R4V
 
 | | Structure of Salt-inducible kinase 3 in complex with inhibitor | | Descriptor: | 1-(2,4-dimethoxyphenyl)-3-(2,6-dimethylphenyl)-1-[6-[[4-(4-methylpiperazin-1-yl)phenyl]amino]pyrimidin-4-yl]urea, Serine/threonine-protein kinase SIK3 | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2023-11-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structures of salt-inducible kinase 3 in complex with inhibitors reveal determinants for binding and selectivity.
J.Biol.Chem., 300, 2024
|
|
8R4O
 
 | | Salt inducible kinase 3 in complex with inhibitor | | Descriptor: | 2-[bis(fluoranyl)methoxy]-4-[6-(2-cyanopropan-2-yl)pyrazolo[1,5-a]pyridin-3-yl]-~{N}-[(1~{R},2~{S})-2-fluoranylcyclopropyl]-6-methoxy-benzamide, SULFATE ION, Serine/threonine-protein kinase SIK3, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2023-11-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.725 Å) | | Cite: | The structures of salt-inducible kinase 3 in complex with inhibitors reveal determinants for binding and selectivity.
J.Biol.Chem., 300, 2024
|
|
