5MGN
 
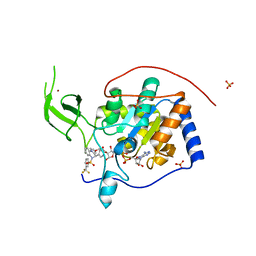 | | Human Sirt6 in complex with activator UBCS38 | | Descriptor: | (4~{R})-4-pyridin-3-yl-5-[3-(trifluoromethyl)phenyl]sulfonyl-4~{H}-pyrrolo[1,2-a]quinoxaline, 1,2-ETHANEDIOL, NAD-dependent protein deacetylase sirtuin-6, ... | | Authors: | Steegborn, C, You, W, Kambach, C. | | Deposit date: | 2016-11-21 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural Basis of Sirtuin 6 Activation by Synthetic Small Molecules.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6HOY
 
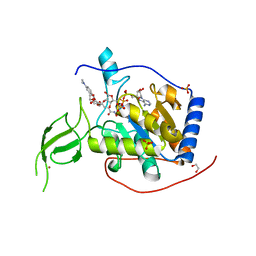 | |
3ZGV
 
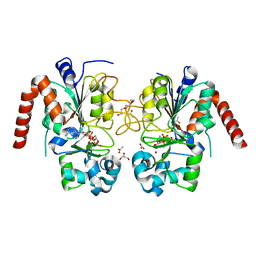 | | Structure of human SIRT2 in complex with ADP-ribose | | Descriptor: | ACETATE ION, GLYCEROL, NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-2, ... | | Authors: | Moniot, S, Steegborn, C. | | Deposit date: | 2012-12-19 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure Analysis of Human Sirt2 and its Adp-Ribose Complex
J.Struct.Biol., 182, 2013
|
|
6L65
 
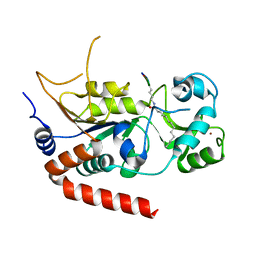 | | Sirtuin 2 protein with H3K18 myristoylated peptide | | Descriptor: | MYRISTIC ACID, NAD-dependent protein deacetylase sirtuin-2, PRO-ARG-LYS-GLN-LEU, ... | | Authors: | Chen, L.F. | | Deposit date: | 2019-10-28 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sirtuin 2 protein with H3K18 myristoylated peptide
To Be Published
|
|
4BN5
 
 | | Structure of human SIRT3 in complex with SRT1720 inhibitor | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, GLYCEROL, N-{2-[3-(piperazin-1-ylmethyl)imidazo[2,1-b][1,3]thiazol-6-yl]phenyl}quinoxaline-2-carboxamide, ... | | Authors: | Nguyen, G.T.T, Schaefer, S, Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-05-13 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structures of Human Sirtuin 3 Complexes with Adp-Ribose and with Carba-Nad+ and Srt1720: Binding Details and Inhibition Mechanism
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6LJM
 
 | |
4BVB
 
 | | CRYSTAL STRUCTURE OF HUMAN SIRT3 IN COMPLEX WITH THE INHIBITOR EX-527 AND ADP-RIBOSE | | Descriptor: | (1S)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1- carboxamide, NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-3, MITOCHONDRIAL, ... | | Authors: | Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-25 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6L66
 
 | |
6LJN
 
 | |
4BN4
 
 | | Structure of human SIRT3 in complex with ADP-ribose | | Descriptor: | 2-[2-[2-[2-[2-(2-hydroxyethyloxy)ethoxy]ethoxy]ethoxy]ethoxy]ethanoic acid, NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-3, MITOCHONDRIAL, ... | | Authors: | Nguyen, G.T.T, Schaefer, S, Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-05-13 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of Human Sirtuin 3 Complexes with Adp-Ribose and with Carba-Nad+ and Srt1720: Binding Details and Inhibition Mechanism
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2QQF
 
 | | Hst2 bound to ADP-HPD and Acetylated histone H4 | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, Histone H4, NAD-dependent deacetylase HST2, ... | | Authors: | Marmorstein, R, Sanders, B.D, Zhao, K, Slama, J. | | Deposit date: | 2007-07-26 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for nicotinamide inhibition and base exchange in sir2 enzymes.
Mol.Cell, 25, 2007
|
|
2QQG
 
 | | Hst2 bound to ADP-HPD, acetyllated histone H4 and nicotinamide | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, Histone H4, NAD-dependent deacetylase HST2, ... | | Authors: | Marmorstein, R, Sanders, B, Zhao, K, Slama, J. | | Deposit date: | 2007-07-26 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for nicotinamide inhibition and base exchange in sir2 enzymes.
Mol.Cell, 25, 2007
|
|
4R8M
 
 | | Human SIRT2 crystal structure in complex with BHJH-TM1 | | Descriptor: | BHJH-TM1 peptide, NAD-dependent protein deacetylase sirtuin-2, ZINC ION, ... | | Authors: | Teng, Y.B, Hao, Q, Lin, H.N, Jing, H. | | Deposit date: | 2014-09-02 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Efficient Demyristoylase Activity of SIRT2 Revealed by Kinetic and Structural Studies
Sci Rep, 5, 2015
|
|
4RMG
 
 | | Human Sirt2 in complex with SirReal2 and NAD+ | | Descriptor: | 2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]-N-[5-(naphthalen-1-ylmethyl)-1,3-thiazol-2-yl]acetamide, NAD-dependent protein deacetylase sirtuin-2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
4RMH
 
 | | Human Sirt2 in complex with SirReal2 and Ac-Lys-H3 peptide | | Descriptor: | 2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]-N-[5-(naphthalen-1-ylmethyl)-1,3-thiazol-2-yl]acetamide, Ac-Lys-H3 peptide, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
4RMJ
 
 | | Human Sirt2 in complex with ADP ribose and nicotinamide | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
4TWI
 
 | | The structure of Sir2Af1 bound to a succinylated histone peptide | | Descriptor: | GLYCEROL, NAD-dependent protein deacylase 1, Succinylated H4 Peptide (aa8-20), ... | | Authors: | Ringel, A.E, Roman, C, Wolberger, C. | | Deposit date: | 2014-06-30 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Alternate deacylating specificities of the archaeal sirtuins Sir2Af1 and Sir2Af2.
Protein Sci., 23, 2014
|
|
6RXK
 
 | | Crystal structure of CobB wt in complex with H4K16-Butyryl peptide | | Descriptor: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXS
 
 | | Crystal structure of CobB Ac3(A76G,Y92A, I131L, V187Y) in complex with H4K16-Acetyl peptide | | Descriptor: | GLYCEROL, Histone H4, NAD-dependent protein deacylase, ... | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXJ
 
 | | Crystal structure of CobB wt in complex with H4K16-Acetyl peptide | | Descriptor: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXL
 
 | | Crystal structure of CobB wt in complex with H4K16-Crotonyl peptide | | Descriptor: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXR
 
 | | Crystal structure of CobB Ac2 (A76G, I131C, V162G) in complex with H4K16Cr-2'OH-ADPr peptide intermediate after co-crystallisation | | Descriptor: | Histone H4, NAD-dependent protein deacylase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{R},4~{R},5~{S})-4-[(~{E})-but-2-enoxy]-3,5-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXP
 
 | | Crystal structure of CobB Ac2 (A76G,I131C,V162A) in complex with H4K16-Crotonyl peptide | | Descriptor: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
4RMI
 
 | | Human Sirt2 in complex with SirReal1 and Ac-Lys-OTC peptide | | Descriptor: | Ac-Lys-OTC peptide, N-(5-benzyl-1,3-thiazol-2-yl)-2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]acetamide, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
6RXO
 
 | | Crystal structure of CobB Ac2 (A76G, I131C, V162A) in complex with H4K16-Buturyl peptide | | Descriptor: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
