1K38
 
 | | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-2 | | Descriptor: | Beta-lactamase OXA-2, FORMIC ACID | | Authors: | Kerff, F, Fonze, E, Bouillenne, F, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-02 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-2
To be Published
|
|
1K39
 
 | | The structure of yeast delta3-delta2-enoyl-COA isomerase complexed with octanoyl-COA | | Descriptor: | OCTANOYL-COENZYME A, PHOSPHATE ION, d3,d2-enoyl CoA isomerase ECI1 | | Authors: | Mursula, A.M, Geerlof, A, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2001-10-02 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: |
|
|
1K3A
 
 | | Structure of the Insulin-like Growth Factor 1 Receptor Kinase | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, insulin receptor substrate 1, insulin-like growth factor 1 receptor | | Authors: | Favelyukis, S, Till, J.H, Hubbard, S.R, Miller, W.T. | | Deposit date: | 2001-10-02 | | Release date: | 2001-11-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and autoregulation of the insulin-like growth factor 1 receptor kinase.
Nat.Struct.Biol., 8, 2001
|
|
1K3B
 
 | | Crystal Structure of Human Dipeptidyl Peptidase I (Cathepsin C): Exclusion Domain Added to an Endopeptidase Framework Creates the Machine for Activation of Granular Serine Proteases | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, SULFATE ION, ... | | Authors: | Turk, D, Janjic, V, Stern, I, Podobnik, M, Lamba, D, Dahl, S.W, Lauritzen, C, Pedersen, J, Turk, V, Turk, B. | | Deposit date: | 2001-10-02 | | Release date: | 2002-04-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of human dipeptidyl peptidase I (cathepsin C): exclusion domain added to an endopeptidase framework creates the machine for activation of granular serine proteases.
EMBO J., 20, 2001
|
|
1K3C
 
 | | Phosphoenolpyruvate carboxykinase in complex with ADP, AlF3 and Pyruvate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Sudom, A.M, Prasad, L, Goldie, H, Delbaere, L.T.J. | | Deposit date: | 2001-10-02 | | Release date: | 2001-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The phosphoryl-transfer mechanism of Escherichia coli phosphoenolpyruvate carboxykinase from the use of AlF(3).
J.Mol.Biol., 314, 2001
|
|
1K3D
 
 | | Phosphoenolpyruvate carboxykinase in complex with ADP and AlF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Sudom, A.M, Prasad, L, Goldie, H, Delbaere, L.T.J. | | Deposit date: | 2001-10-02 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The phosphoryl-transfer mechanism of Escherichia coli phosphoenolpyruvate carboxykinase from the use of AlF(3).
J.Mol.Biol., 314, 2001
|
|
1K3E
 
 | | Type III secretion chaperone CesT | | Descriptor: | CesT | | Authors: | Luo, Y, Bertero, M, Frey, E.A, Pfuetzner, R.A, Wenk, M.R, Creagh, L, Marcus, S.L, Lim, D, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2001-10-02 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical characterization of the type III secretion chaperones CesT and SigE.
Nat.Struct.Biol., 8, 2001
|
|
1K3F
 
 | | Uridine Phosphorylase from E. coli, Refined in the Monoclinic Crystal Lattice | | Descriptor: | uridine phosphorylase | | Authors: | Morgunova, E.Yu, Mikhailov, A.M, Popov, A.N, Blagova, E.V, Smirnova, E.A, Vainshtein, B.K, Mao, C, Armstrong, S.R, Ealick, S.E, Komissarov, A.A, Linkova, E.V, Burlakova, A.A, Mironov, A.S, Debabov, V.G. | | Deposit date: | 2001-10-02 | | Release date: | 2001-10-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic structure at 2.5 A resolution of uridine phosphorylase from E. coli as refined in the monoclinic crystal lattice.
FEBS Lett., 367, 1995
|
|
1K3G
 
 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
1K3H
 
 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
1K3I
 
 | | Crystal Structure of the Precursor of Galactose Oxidase | | Descriptor: | ACETATE ION, CALCIUM ION, Galactose Oxidase Precursor, ... | | Authors: | Firbank, S.J, Rogers, M.S, Wilmot, C.M, Dooley, D.M, Halcrow, M.A, Knowles, P.F, McPherson, M.J, Phillips, S.E.V. | | Deposit date: | 2001-10-03 | | Release date: | 2001-11-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the precursor of galactose oxidase: an unusual self-processing enzyme.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1K3J
 
 | | Refined NMR Structure of the FHA1 Domain of Yeast Rad53 | | Descriptor: | Protein Kinase SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-03 | | Release date: | 2001-12-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
1K3K
 
 | | Solution Structure of a Bcl-2 Homolog from Kaposi's Sarcoma Virus | | Descriptor: | functional anti-apoptotic factor vBCL-2 homolog | | Authors: | Huang, Q, Petros, A.M, Virgin, H.W, Fesik, S.W, Olejniczak, E.T. | | Deposit date: | 2001-10-03 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a Bcl-2 homolog from Kaposi sarcoma virus.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1K3L
 
 | | Crystal Structure Analysis of S-hexyl-glutathione Complex of Glutathione Transferase at 1.5 Angstroms Resolution | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1, S-HEXYLGLUTATHIONE | | Authors: | Le Trong, I, Stenkamp, R.E, Ibarra, C, Atkins, W.M, Adman, E.T. | | Deposit date: | 2001-10-03 | | Release date: | 2002-10-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.3-A resolution structure of human glutathione S-transferase with S-hexyl
glutathione bound reveals possible extended ligandin binding site
Proteins, 48, 2002
|
|
1K3M
 
 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT ILE-A2-ALA, HIS-B10-ASP, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | Descriptor: | INSULIN | | Authors: | Xu, B, Hua, Q.-X, Nakagawa, S.H, Jia, W, Chu, Y.-C, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-17 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | A cavity-forming mutation in insulin induces segmental unfolding of a surrounding alpha-helix.
Protein Sci., 11, 2002
|
|
1K3N
 
 | | NMR Structure of the FHA1 Domain of Rad53 in Complex with a Rad9-derived Phosphothreonine (at T155) Peptide | | Descriptor: | DNA repair protein Rad9, Protein Kinase SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-03 | | Release date: | 2001-12-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
1K3O
 
 | | Crystal Structure Analysis of apo Glutathione S-Transferase | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1 | | Authors: | Le Trong, I, Stenkamp, R.E, Ibarra, C, Atkins, W.M, Adman, E.T. | | Deposit date: | 2001-10-03 | | Release date: | 2002-10-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.3-A resolution structure of human glutathione S-transferase with S-hexyl glutathione bound reveals possible extended ligandin binding site.
Proteins, 48, 2002
|
|
1K3Q
 
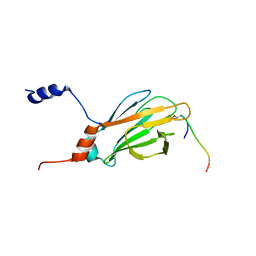 | | NMR structure of the FHA1 Domain of Rad53 in Complex with a Rad9-derived Phosphothreonine (at T192) Peptide | | Descriptor: | DNA repair protein Rad9, Protein Kinase SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-03 | | Release date: | 2001-12-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
1K3R
 
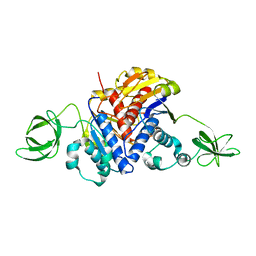 | | Crystal Structure of the Methyltransferase with a Knot from Methanobacterium thermoautotrophicum | | Descriptor: | conserved protein MT0001 | | Authors: | Zarembinski, T.I, Kim, Y, Peterson, K, Christendat, D, Dharamsi, A, Arrowsmith, C.H, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-03 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Deep trefoil knot implicated in RNA binding found in an archaebacterial protein.
Proteins, 50, 2003
|
|
1K3S
 
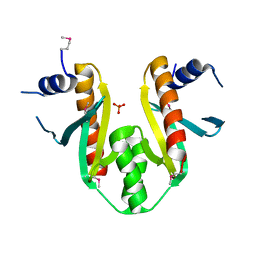 | | Type III Secretion Chaperone SigE | | Descriptor: | PHOSPHATE ION, SigE | | Authors: | Bertero, M.G, Luo, Y, Frey, E.A, Pfuetzner, R.A, Wenk, M.R, Creagh, L, Marcus, S.L, Lim, D, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2001-10-03 | | Release date: | 2001-11-28 | | Last modified: | 2016-05-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical characterization of the type III secretion chaperones CesT and SigE.
Nat.Struct.Biol., 8, 2001
|
|
1K3T
 
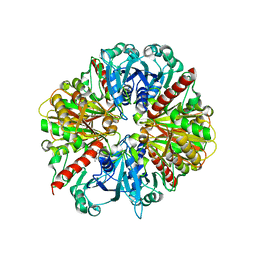 | | Structure of Glycosomal Glyceraldehyde-3-Phosphate Dehydrogenase from Trypanosoma cruzi Complexed with Chalepin, a Coumarin Derivative Inhibitor | | Descriptor: | 6-(1,1-DIMETHYLALLYL)-2-(1-HYDROXY-1-METHYLETHYL)-2,3-DIHYDRO-7H-FURO[3,2-G]CHROMEN-7-ONE, Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Pavao, F. | | Deposit date: | 2001-10-04 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Trypanosoma cruzi glycosomal glyceraldehyde-3-phosphate dehydrogenase complexed with chalepin, a natural product inhibitor, at 1.95 A resolution.
FEBS Lett., 520, 2002
|
|
1K3U
 
 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE COMPLEXED WITH N-[1H-INDOL-3-YL-ACETYL]ASPARTIC ACID | | Descriptor: | N-[1H-INDOL-3-YL-ACETYL]ASPARTIC ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | Deposit date: | 2001-10-04 | | Release date: | 2002-07-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of a new class of allosteric effectors complexed to tryptophan synthase.
J.Biol.Chem., 277, 2002
|
|
1K3V
 
 | | Porcine Parvovirus Capsid | | Descriptor: | capsid protein VP2 | | Authors: | Simpson, A.A, Hebert, B, Sullivan, G.M, Parrish, C.R, Zadori, Z, Tijssen, P, Rossmann, M.G. | | Deposit date: | 2001-10-04 | | Release date: | 2001-10-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of porcine parvovirus: comparison with related viruses.
J.Mol.Biol., 315, 2002
|
|
1K3W
 
 | | Crystal structure of a trapped reaction intermediate of the DNA Repair Enzyme Endonuclease VIII with DNA | | Descriptor: | 5'-D(*CP*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3', 5'-D(*GP*GP*CP*TP*TP*CP*AP*TP*CP*CP*TP*GP*G)-3', Endonuclease VIII, ... | | Authors: | Golan, G, Zharkov, D.O, Gilboa, R, Fernandes, A.S, Kycia, J.H, Gerchman, S.E, Rieger, R.A, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-10-04 | | Release date: | 2002-10-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate.
EMBO J., 21, 2002
|
|
1K3X
 
 | | Crystal structure of a trapped reaction intermediate of the DNA repair enzyme Endonuclease VIII with Brominated-DNA | | Descriptor: | 5'-D(*CP*CP*AP*GP*GP*AP*(PED)P*GP*AP*AP*GP*CP*C)-3', 5'-D(*GP*GP*CP*(BRU)P*(BRU)P*CP*AP*(BRU)P*CP*CP*(BRU)P*GP*G)-3', Endonuclease VIII, ... | | Authors: | Golan, G, Zharkov, D.O, Gilboa, R, Fernandes, A.S, Kycia, J.H, Gerchman, S.E, Rieger, R.A, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-10-04 | | Release date: | 2002-10-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate.
EMBO J., 21, 2002
|
|
