3PRL
 
 | | Crystal structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Bacillus halodurans C-125 | | 分子名称: | NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | 著者 | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2010-11-29 | | 公開日 | 2010-12-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Bacillus halodurans C-125
To be Published
|
|
7RN5
 
 | |
3C0K
 
 | |
6SNE
 
 | | crystal structure of LN01 Fab in complex with an HIV-1 gp41 peptide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, LN01 heavy chain, ... | | 著者 | Caillat, C, Pinto, D, Corti, D, Fenwick, C, Pantaleo, G, Weissenhorn, W. | | 登録日 | 2019-08-23 | | 公開日 | 2019-11-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.9 Å) | | 主引用文献 | Structural Basis for Broad HIV-1 Neutralization by the MPER-Specific Human Broadly Neutralizing Antibody LN01.
Cell Host Microbe, 26, 2019
|
|
3PT5
 
 | | Crystal structure of NanS | | 分子名称: | NANS (YJHS), A 9-O-acetyl N-acetylneuraminic acid esterase | | 著者 | Ruane, K.M, Rangarajan, E.S, Proteau, A, Schrag, J.D, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | 登録日 | 2010-12-02 | | 公開日 | 2011-05-18 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural and enzymatic characterization of NanS (YjhS), a 9-O-Acetyl N-acetylneuraminic acid esterase from Escherichia coli O157:H7.
Protein Sci., 20, 2011
|
|
3CCT
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | 分子名称: | (3R,5R)-7-[2-(4-fluorophenyl)-4-[(2-hydroxyphenyl)carbamoyl]-5-(1-methylethyl)-3-phenyl-1H-pyrrol-1-yl]-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | 著者 | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | 登録日 | 2008-02-26 | | 公開日 | 2008-06-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
3CCZ
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | 分子名称: | (3R,5R)-7-[2-(4-fluorophenyl)-4-{[(1S)-2-hydroxy-1-phenylethyl]carbamoyl}-5-(1-methylethyl)-1H-imidazol-1-yl]-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, SULFATE ION | | 著者 | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | 登録日 | 2008-02-26 | | 公開日 | 2008-06-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
3CD5
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | 分子名称: | (3R,5R)-7-[3-(biphenyl-4-ylcarbamoyl)-2-ethyl-5,6,7,8-tetrahydrocyclohepta[b]pyrrol-1(4H)-yl]-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, SULFATE ION | | 著者 | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | 登録日 | 2008-02-26 | | 公開日 | 2008-06-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
3PXW
 
 | | Crystal Structure of Ferrous NO Adduct of MauG in Complex with Pre-Methylamine Dehydrogenase | | 分子名称: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ACETATE ION, ... | | 著者 | Yukl, E.T, Goblirsch, B.R, Wilmot, C.M. | | 登録日 | 2010-12-10 | | 公開日 | 2011-03-23 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Crystal Structures of CO and NO Adducts of MauG in Complex with Pre-Methylamine Dehydrogenase: Implications for the Mechanism of Dioxygen Activation.
Biochemistry, 50, 2011
|
|
3C8R
 
 | |
3PZL
 
 | |
6BVD
 
 | |
3BND
 
 | | Lipoxygenase-1 (Soybean), I553V Mutant | | 分子名称: | FE (III) ION, Seed lipoxygenase-1 | | 著者 | Tomchick, D.R. | | 登録日 | 2007-12-14 | | 公開日 | 2008-04-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Enzyme structure and dynamics affect hydrogen tunneling: the impact of a remote side chain (I553) in soybean lipoxygenase-1.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5OP9
 
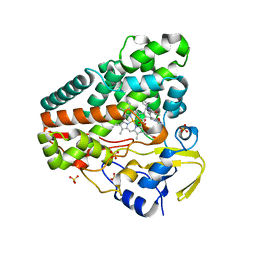 | | The crystal structure of P450 CYP121 in complex with lead compound 7e | | 分子名称: | 4-(imidazol-1-ylmethyl)-3-(4-methoxyphenyl)-1-phenyl-pyrazole, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Levy, C.W. | | 登録日 | 2017-08-09 | | 公開日 | 2018-03-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.455 Å) | | 主引用文献 | Novel Aryl Substituted Pyrazoles as Small Molecule Inhibitors of Cytochrome P450 CYP121A1: Synthesis and Antimycobacterial Evaluation.
J. Med. Chem., 60, 2017
|
|
3CAQ
 
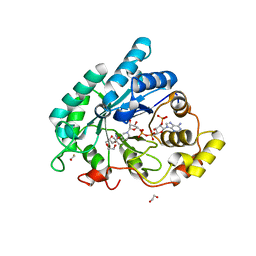 | | Crystal structure of 5beta-reductase (AKR1D1) in complex with NADPH | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3-oxo-5-beta-steroid 4-dehydrogenase, ... | | 著者 | Faucher, F, Cantin, L, Breton, R. | | 登録日 | 2008-02-20 | | 公開日 | 2008-12-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structures of Human Delta4-3-Ketosteroid 5beta-Reductase (AKR1D1) Reveal the Presence of an Alternative Binding Site Responsible for Substrate Inhibition (dagger) (,) (double dagger).
Biochemistry, 47, 2008
|
|
1O6L
 
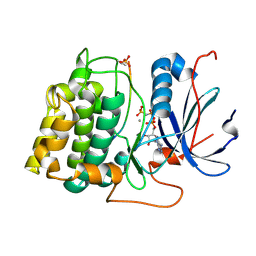 | | Crystal structure of an activated Akt/protein kinase B (PKB-PIF chimera) ternary complex with AMP-PNP and GSK3 peptide | | 分子名称: | GLYCOGEN SYNTHASE KINASE-3 BETA, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Yang, J, Cron, P, Good, V.M, Thompson, V, Hemmings, B.A, Barford, D. | | 登録日 | 2002-10-08 | | 公開日 | 2002-11-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structure of an Activated Akt/Protein Kinase B Ternary Complex with Gsk-3 Peptide and AMP-Pnp
Nat.Struct.Biol., 9, 2002
|
|
1O6K
 
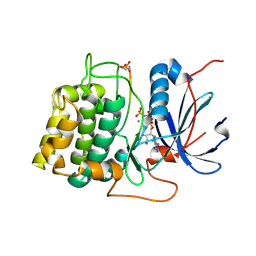 | | Structure of activated form of PKB kinase domain S474D with GSK3 peptide and AMP-PNP | | 分子名称: | GLYCOGEN SYNTHASE KINASE-3 BETA, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Yang, J, Cron, P, Good, V.M, Thompson, V, Hemmings, B.A, Barford, D. | | 登録日 | 2002-10-08 | | 公開日 | 2002-11-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal Structure of an Activated Akt/Protein Kinase B Ternary Complex with Gsk-3 Peptide and AMP-Pnp
Nat.Struct.Biol., 9, 2002
|
|
3PTQ
 
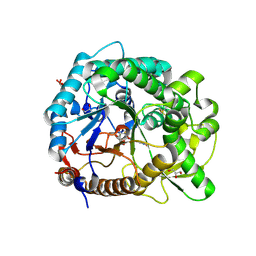 | | The crystal structure of rice (Oryza sativa L.) Os4BGlu12 with dinitrophenyl 2-deoxy-2-fluoro-beta-D-glucopyranoside | | 分子名称: | 2,4-dinitrophenyl 2-deoxy-2-fluoro-beta-D-glucopyranoside, Beta-glucosidase Os4BGlu12, GLYCEROL, ... | | 著者 | Sansenya, S, Opassiri, R, Kuaprasert, B, Chen, C.J, Ketudat Cairns, J.R. | | 登録日 | 2010-12-03 | | 公開日 | 2011-05-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | The crystal structure of rice (Oryza sativa L.) Os4BGlu12, an oligosaccharide and tuberonic acid glucoside-hydrolyzing beta-glucosidase with significant thioglucohydrolase activity
Arch.Biochem.Biophys., 510, 2011
|
|
6TCH
 
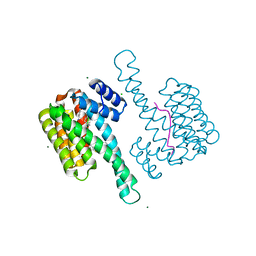 | |
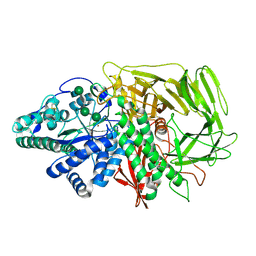
3BVX
 
 | |
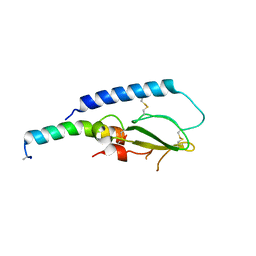
5OTV
 
 | |
5OQM
 
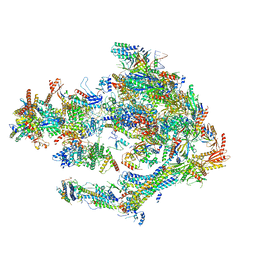 | | STRUCTURE OF YEAST TRANSCRIPTION PRE-INITIATION COMPLEX WITH TFIIH AND CORE MEDIATOR | | 分子名称: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | 著者 | Schilbach, S, Hantsche, M, Tegunov, D, Dienemann, C, Wigge, C, Urlaub, H, Cramer, P. | | 登録日 | 2017-08-13 | | 公開日 | 2018-05-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (5.8 Å) | | 主引用文献 | Structures of transcription pre-initiation complex with TFIIH and Mediator.
Nature, 551, 2017
|
|
3PNM
 
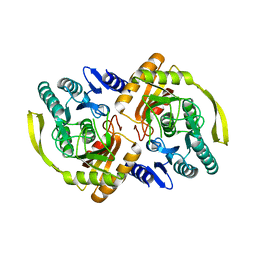 | | Crystal Structure of E.coli Dha kinase DhaK (H56A) | | 分子名称: | PTS-dependent dihydroxyacetone kinase, dihydroxyacetone-binding subunit dhaK | | 著者 | Shi, R, McDonald, L, Matte, A, Cygler, M, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | 登録日 | 2010-11-19 | | 公開日 | 2011-01-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural and mechanistic insight into covalent substrate binding by Escherichia coli dihydroxyacetone kinase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PZB
 
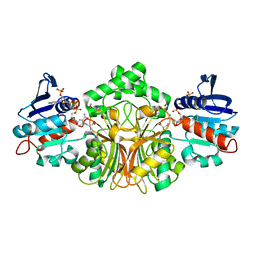 | | Crystals Structure of Aspartate beta-Semialdehyde Dehydrogenase complex with NADP and D-2,3-Diaminopropionate | | 分子名称: | 1,2-ETHANEDIOL, 3-amino-D-alanine, Aspartate-semialdehyde dehydrogenase, ... | | 著者 | Pavlovsky, A.G, Viola, R.E. | | 登録日 | 2010-12-14 | | 公開日 | 2012-01-04 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Characterization of Inhibitors with Selectivity against Members of a Homologous Enzyme Family.
Chem.Biol.Drug Des., 79, 2012
|
|
6CAY
 
 | |
