6BN0
 
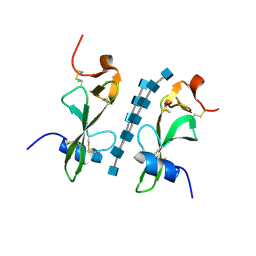 | | Avirulence protein 4 (Avr4) from Cladosporium fulvum bound to the hexasaccharide of chitin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Race-specific elicitor A4 | | 著者 | Hurlburt, N.K, Fisher, A.J. | | 登録日 | 2017-11-15 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure of the Cladosporium fulvum Avr4 effector in complex with (GlcNAc)6 reveals the ligand-binding mechanism and uncouples its intrinsic function from recognition by the Cf-4 resistance protein.
PLoS Pathog., 14, 2018
|
|
6I2Y
 
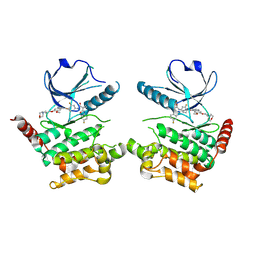 | | Human STK10 bound to Foretinib | | 分子名称: | N-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide, Serine/threonine-protein kinase 10 | | 著者 | Sorrell, F.J, Berger, B.-T, Oerum, S, von Delft, F, Bountra, C, Arrowsmith, C, Edwards, A.M, Knapp, S, Elkins, J.M. | | 登録日 | 2018-11-02 | | 公開日 | 2018-12-12 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Human STK10 bound to GW683134
To Be Published
|
|
6BNE
 
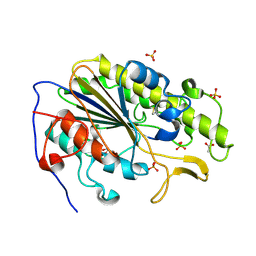 | | Crystal structure of the intrinsic colistin resistance enzyme ICR(Mc) from Moraxella catarrhalis, catalytic domain, phosphate-bound complex | | 分子名称: | ACETATE ION, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Di Leo, R, Savchenko, A, Anderson, W.F, Satchell, K.J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-11-16 | | 公開日 | 2018-01-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Substrate recognition by a colistin resistance enzyme from Moraxella catarrhalis.
ACS Chem. Biol., 2018
|
|
7LCY
 
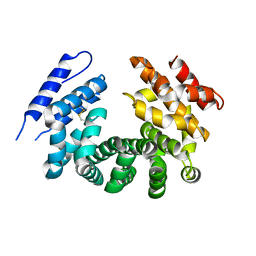 | | Crystal structure of the ligand-free ARM domain from Drosophila SARM1 | | 分子名称: | Isoform B of NAD(+) hydrolase sarm1 | | 著者 | Gu, W, Nanson, J.D, Luo, Z, McGuinness, H.Y, Manik, M.K, Jia, X, Ve, T, Kobe, B. | | 登録日 | 2021-01-12 | | 公開日 | 2021-03-10 | | 最終更新日 | 2021-04-21 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | SARM1 is a metabolic sensor activated by an increased NMN/NAD + ratio to trigger axon degeneration.
Neuron, 109, 2021
|
|
6ZT4
 
 | | Pentapeptide repeat protein MfpA from Mycobacterium smegmatis | | 分子名称: | 1,2-ETHANEDIOL, Pentapeptide repeat protein MfpA | | 著者 | Feng, L, Mundy, J.E.A, Stevenson, C.E.M, Mitchenall, L.A, Lawson, D.M, Mi, K, Maxwell, A. | | 登録日 | 2020-07-17 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | The pentapeptide-repeat protein, MfpA, interacts with mycobacterial DNA gyrase as a DNA T-segment mimic.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8RII
 
 | |
6BNJ
 
 | | Human hypoxanthine guanine phosphoribosyltransferase in complex with [3R,4R]-4-guanin-9-yl-3-((R)-2-hydroxy-2-phosphonoethyl)oxy-1-N-(phosphonopropionyl)pyrrolidine | | 分子名称: | (3-{(3R,4R)-3-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-4-[(2R)-2-hydroxy-2-phosphonoethoxy]pyrrolidin-1-yl}-3-oxopropy l)phosphonic acid, Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION | | 著者 | Keough, D.T, Rejman, D, Guddat, L.W. | | 登録日 | 2017-11-16 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.909 Å) | | 主引用文献 | Design of Plasmodium vivax Hypoxanthine-Guanine Phosphoribosyltransferase Inhibitors as Potential Antimalarial Therapeutics.
ACS Chem. Biol., 13, 2018
|
|
7LVL
 
 | | Dihydrodipicolinate synthase bound with allosteric inhibitor (S)-lysine from Candidatus Liberibacter solanacearum | | 分子名称: | 4-hydroxy-tetrahydrodipicolinate synthase, LYSINE | | 著者 | Gilkes, J.M, Frampton, R.A, Board, A.J, Sheen, C.R, Smith, G.R, Dobson, R.C.J.D. | | 登録日 | 2021-02-25 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Dihydrodipicolinate synthase bound with allosteric inhibitor (S)-lysine from Candidatus Liberibacter solanacearum
To Be Published
|
|
7Q9Q
 
 | |
6BMC
 
 | | The structure of a dimeric type II DAH7PS associated with pyocyanin biosynthesis in Pseudomonas aeruginosa | | 分子名称: | CHLORIDE ION, COBALT (II) ION, PHOSPHOENOLPYRUVATE, ... | | 著者 | Sterritt, O.W, Jameson, G.B, Parker, E.J. | | 登録日 | 2017-11-14 | | 公開日 | 2018-10-03 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural and functional characterisation of the entry point to pyocyanin biosynthesis inPseudomonas aeruginosadefines a new 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase subclass.
Biosci. Rep., 38, 2018
|
|
6I6K
 
 | | Papaver somniferum O-methyltransferase 1 | | 分子名称: | (13aS)-3,10-dimethoxy-5,8,13,13a-tetrahydro-6H-isoquino[3,2-a]isoquinoline-2,9-diol, O-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Cabry, M.P, Offen, W.A, Winzer, T, Li, Y, Graham, I.A, Davies, G.J, Saleh, P. | | 登録日 | 2018-11-15 | | 公開日 | 2019-03-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Structure of Papaver somniferum O-Methyltransferase 1 Reveals Initiation of Noscapine Biosynthesis with Implications for Plant Natural Product Methylation
Acs Catalysis, 2019
|
|
6BMQ
 
 | | Crystal structure of Arabidopsis Dehydroquinate dehydratase-shikimate dehydrogenase (T381G mutant) in complex with tartrate and shikimate | | 分子名称: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic, ... | | 著者 | Christendat, D, Peek, J. | | 登録日 | 2017-11-15 | | 公開日 | 2018-09-26 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.077 Å) | | 主引用文献 | Structural and biochemical approaches uncover multiple evolutionary trajectories of plant quinate dehydrogenases.
Plant J., 2018
|
|
7L6W
 
 | | SFX structure of the MyD88 TIR domain higher-order assembly | | 分子名称: | Myeloid differentiation primary response protein MyD88 | | 著者 | Clabbers, M.T.B, Holmes, S, Muusse, T.W, Vajjhala, P, Thygesen, S.J, Malde, A.K, Hunter, D.J.B, Croll, T.I, Flueckiger, L, Nanson, J.D, Rahaman, M.H, Aquila, A, Hunter, M.S, Liang, M, Yoon, C.H, Zhao, J, Zatsepin, N.A, Abbey, B, Sierecki, E, Gambin, Y, Stacey, K.J, Darmanin, C, Kobe, B, Xu, H, Ve, T. | | 登録日 | 2020-12-24 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | MyD88 TIR domain higher-order assembly interactions revealed by microcrystal electron diffraction and serial femtosecond crystallography.
Nat Commun, 12, 2021
|
|
7A6O
 
 | |
6I3B
 
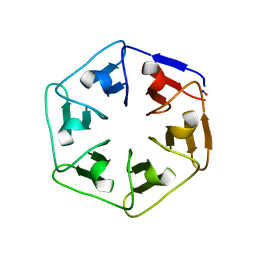 | |
7LCZ
 
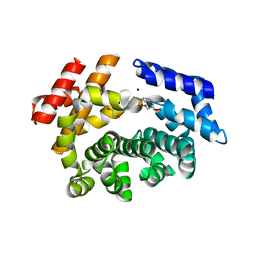 | | Crystal structure of the ARM domain from Drosophila SARM1 in complex with NMN | | 分子名称: | 1,2-ETHANEDIOL, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Isoform B of NAD(+) hydrolase sarm1, ... | | 著者 | Gu, W, Nanson, J.D, Luo, Z, Jia, X, Manik, M.K, Ve, T, Kobe, B. | | 登録日 | 2021-01-12 | | 公開日 | 2021-03-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | SARM1 is a metabolic sensor activated by an increased NMN/NAD + ratio to trigger axon degeneration.
Neuron, 109, 2021
|
|
6ZT3
 
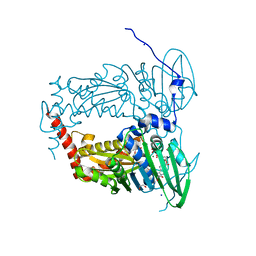 | | N-terminal 47 kDa fragment of the Mycobacterium smegmatis DNA Gyrase B subunit complexed with ADPNP | | 分子名称: | 1,2-ETHANEDIOL, DNA gyrase subunit B, MAGNESIUM ION, ... | | 著者 | Feng, L, Mundy, J.E.A, Stevenson, C.E.M, Mitchenall, L.A, Lawson, D.M, Mi, K, Maxwell, A. | | 登録日 | 2020-07-17 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | The pentapeptide-repeat protein, MfpA, interacts with mycobacterial DNA gyrase as a DNA T-segment mimic.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8RG2
 
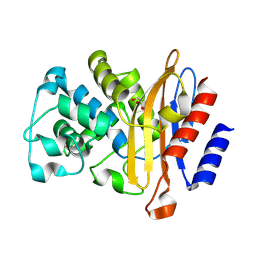 | |
6I9C
 
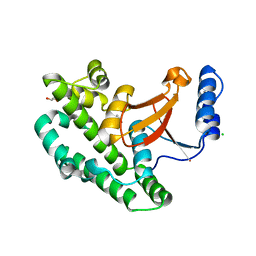 | | Structure of the OTU domain of OTULIN G281R mutant | | 分子名称: | CHLORIDE ION, GLYCEROL, Ubiquitin thioesterase otulin | | 著者 | Damgaard, R.B, Elliott, P.R, Swatek, K.N, Maher, E.R, Stepensky, P, Elpeleg, O, Komander, D, Berkun, Y. | | 登録日 | 2018-11-22 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | OTULIN deficiency in ORAS causes cell type-specific LUBAC degradation, dysregulated TNF signalling and cell death.
Embo Mol Med, 11, 2019
|
|
7A6X
 
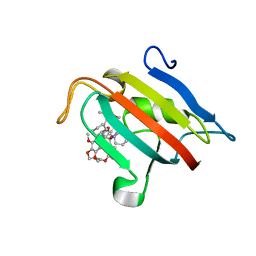 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 56 | | 分子名称: | (2S,9S,12R)-2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-24,27-dimethoxy-11,18,22-trioxa-4-azatetracyclo[21.2.2.113,17.04,9]octacosa-1(25),13(28),14,16,23,26-hexaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | 著者 | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | 登録日 | 2020-08-27 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
8RD5
 
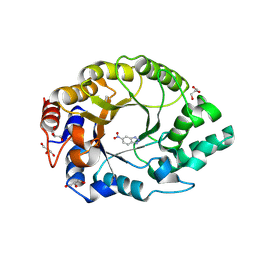 | | Crystal structure of Kemp Eliminase HG3.R5 with bound transition state analog 6-nitrobenzotriazole | | 分子名称: | 6-NITROBENZOTRIAZOLE, ACETATE ION, Endo-1,4-beta-xylanase, ... | | 著者 | Schaub, D, Schwander, T, Hueppi, S, Buller, R.M. | | 登録日 | 2023-12-07 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Enriching productive mutational paths accelerates enzyme evolution.
Nat.Chem.Biol., 2024
|
|
6BPM
 
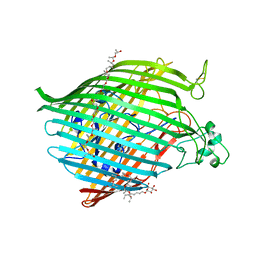 | | The crystal structure of the Ferric-Catecholate import receptor Fiu from K12 E. coli: Closed form (C21) | | 分子名称: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, Catecholate siderophore receptor Fiu, octyl beta-D-glucopyranoside | | 著者 | Grinter, R. | | 登録日 | 2017-11-23 | | 公開日 | 2018-11-28 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The structure of the bacterial iron-catecholate transporter Fiu suggests that it imports substrates via a two-step mechanism.
J.Biol.Chem., 2019
|
|
7A6P
 
 | |
7AD6
 
 | | Crystal structure of human complement C5 in complex with the K92 bovine knob domain peptide. | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CYSTEINE, ... | | 著者 | Macpherson, A, van den Elsen, J.M.H, Schulze, M.E, Birtley, J.R. | | 登録日 | 2020-09-14 | | 公開日 | 2021-03-10 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | The allosteric modulation of Complement C5 by knob domain peptides.
Elife, 10, 2021
|
|
6BHI
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | 分子名称: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, UNKNOWN ATOM OR ION | | 著者 | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2017-10-30 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
