7DQJ
 
 | | E. coli GyrB ATPase domain in complex with 3,4-Dihydroxyacetophenone | | 分子名称: | 1-[3,4-bis(oxidanyl)phenyl]ethanone, 1H-benzimidazol-2-amine, DNA gyrase subunit B, ... | | 著者 | Yu, Y, Zhou, H. | | 登録日 | 2020-12-24 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Identification of new building blocks by fragment screening for discovering GyrB inhibitors.
Bioorg.Chem., 114, 2021
|
|
7DQF
 
 | |
7DQU
 
 | |
7DQL
 
 | | E. coli GyrB ATPase domain in complex with 4-chlorobenzene-1,2-diol | | 分子名称: | 1H-benzimidazol-2-amine, 4-CHLOROBENZENE-1,2-DIOL, DNA gyrase subunit B, ... | | 著者 | Yu, Y, Zhou, H. | | 登録日 | 2020-12-24 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Identification of new building blocks by fragment screening for discovering GyrB inhibitors.
Bioorg.Chem., 114, 2021
|
|
7DQM
 
 | | E. coli GyrB ATPase domain in complex with Naringenin | | 分子名称: | 1H-benzimidazol-2-amine, DNA gyrase subunit B, PHOSPHATE ION, ... | | 著者 | Yu, Y, Zhou, H. | | 登録日 | 2020-12-24 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Identification of new building blocks by fragment screening for discovering GyrB inhibitors.
Bioorg.Chem., 114, 2021
|
|
7DQS
 
 | |
7DQW
 
 | |
3ZXQ
 
 | |
4ASG
 
 | | The structure of modified benzoquinone ansamycins bound to yeast N- terminal Hsp90 | | 分子名称: | ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, NICKEL (II) ION, [(3R,5S,6R,7R,11S,12Z,14E)-5,11,21-trimethoxy-3,7,9,15-tetramethyl-6-oxidanyl-16,20,22-tris(oxidanylidene)-19-phenyl-17-azabicyclo[16.3.1]docosa-1(21),8,12,14,18-pentaen-10-yl] carbamate | | 著者 | Roe, S.M, Prodromou, C. | | 登録日 | 2012-05-01 | | 公開日 | 2013-04-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Synthesis of 19-Substituted Geldanamycins with Altered Conformations and Their Binding to Heat Shock Protein Hsp90.
Nat.Chem., 5, 2013
|
|
4AS9
 
 | | The structure of modified benzoquinone ansamycins bound to yeast N- terminal Hsp90 | | 分子名称: | ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, [(3R,5S,6R,7R,10R,11S,12E)-5,11,21-trimethoxy-3,7,9,15,19-pentamethyl-6-oxidanyl-16,20,22-tris(oxidanylidene)-17-azabicyclo[16.3.1]docosa-1(21),8,12,14,18-pentaen-10-yl] carbamate | | 著者 | Roe, S.M, Prodromou, C. | | 登録日 | 2012-04-30 | | 公開日 | 2013-04-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Synthesis of 19-substituted geldanamycins with altered conformations and their binding to heat shock protein Hsp90.
Nat Chem, 5, 2013
|
|
4ASF
 
 | | The structure of modified benzoquinone ansamycins bound to yeast N- terminal Hsp90 | | 分子名称: | (8S,9R,13R,14S,16R)-21-(furan-2-yl)-13-hydroxy-8,14,19-trimethoxy-16-methyl-4,10,12-trimethylidene-3,20,22-trioxo-2-azabicyclo[16.3.1]docosa-1(21),18-dien-9-yl carbamate, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, NICKEL (II) ION | | 著者 | Roe, S.M, Prodromou, C. | | 登録日 | 2012-05-01 | | 公開日 | 2013-04-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Synthesis of 19-substituted geldanamycins with altered conformations and their binding to heat shock protein Hsp90.
Nat Chem, 5, 2013
|
|
4B6C
 
 | | Structure of the M. smegmatis GyrB ATPase domain in complex with an aminopyrazinamide | | 分子名称: | 6-(3,4-dimethylphenyl)-3-[[4-[3-(4-methylpiperazin-1-yl)propoxy]phenyl]amino]pyrazine-2-carboxamide, DNA gyrase subunit B,DNA gyrase subunit B,DNA gyrase subunit B, SODIUM ION | | 著者 | Tucker, J.A, Shirude, P.S, Madhavapeddi, P, Hussein, S, Basu, R, Ghorpade, S. | | 登録日 | 2012-08-09 | | 公開日 | 2013-01-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Aminopyrazinamides: novel and specific GyrB inhibitors that kill replicating and nonreplicating Mycobacterium tuberculosis.
ACS Chem. Biol., 8, 2013
|
|
4ASA
 
 | | The structure of modified benzoquinone ansamycins bound to yeast N- terminal Hsp90 | | 分子名称: | ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, CHLORIDE ION, [(3R,5S,6R,7R,12E)-5,11-dimethoxy-3,7,9,15,19-pentamethyl-6-oxidanyl-16,20,22-tris(oxidanylidene)-21-(prop-2-enylamino)-17-azabicyclo[16.3.1]docosa-1(21),8,12,14,18-pentaen-10-yl] carbamate | | 著者 | Roe, S.M, Prodromou, C. | | 登録日 | 2012-04-30 | | 公開日 | 2013-04-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Synthesis of 19-Substituted Geldanamycins with Altered Conformations and Their Binding to Heat Shock Protein Hsp90.
Nat.Chem., 5, 2013
|
|
4ASB
 
 | | The structure of modified benzoquinone ansamycins bound to yeast N- terminal Hsp90 | | 分子名称: | (4E,6E,8S,9R,10E,12R,13R,14S,16R)-19-{[2-(dimethylamino)ethyl]amino}-13-hydroxy-8,14-dimethoxy-4,10,12,16,21-pentamethyl-3,20,22-trioxo-2-azabicyclo[16.3.1]docosa-1(21),4,6,10,18-pentaen-9-yl carbamate, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | 著者 | Roe, S.M, Prodromou, C. | | 登録日 | 2012-04-30 | | 公開日 | 2013-04-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (3.08 Å) | | 主引用文献 | Synthesis of 19-substituted geldanamycins with altered conformations and their binding to heat shock protein Hsp90.
Nat Chem, 5, 2013
|
|
4BAE
 
 | | Optimisation of pyrroleamides as mycobacterial GyrB ATPase inhibitors: Structure Activity Relationship and in vivo efficacy in the mouse model of tuberculosis | | 分子名称: | 2-[(3S,4R)-4-[(3-bromanyl-4-chloranyl-5-methyl-1H-pyrrol-2-yl)carbonylamino]-3-methoxy-piperidin-1-yl]-4-(2-methyl-1,2,4-triazol-3-yl)-1,3-thiazole-5-carboxylic acid, CALCIUM ION, DNA GYRASE SUBUNIT B, ... | | 著者 | Read, J.A, Gingell, H.G, Madhavapeddi, P. | | 登録日 | 2012-09-13 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Optimization of Pyrrolamides as Mycobacterial Gyrb ATPase Inhibitors: Structure Activity Relationship and in Vivo Efficacy in the Mouse Model of Tuberculosis.
Antimicrob.Agents Chemother., 58, 2014
|
|
4CE3
 
 | | Hsp90 N-terminal domain bound to macrolactam analogues of radicicol. | | 分子名称: | 13-Chloro-14,16-dihydroxy-2-methyl-2,3,4,5,9,10-hexahydrobenz[c][1]azacyclotetradecine-1,11(8H,12H)-dione, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | 著者 | Roe, S.M, Parry-Morris, S, Prodromou, C. | | 登録日 | 2013-11-08 | | 公開日 | 2014-01-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Synthesis of Macrolactam Analogues of Radicicol and Their Binding to Heat Shock Protein Hsp90.
Org.Biomol.Chem., 12, 2014
|
|
3ZXO
 
 | | CRYSTAL STRUCTURE OF THE MUTANT ATP-BINDING DOMAIN OF MYCOBACTERIUM TUBERCULOSIS DOSS | | 分子名称: | ACETATE ION, GLYCEROL, REDOX SENSOR HISTIDINE KINASE RESPONSE REGULATOR DEVS, ... | | 著者 | Cho, H.Y, Cho, H.J, Kang, B.S. | | 登録日 | 2011-08-13 | | 公開日 | 2011-08-24 | | 最終更新日 | 2025-04-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Activation of ATP Binding for the Autophosphorylation of Doss, a Mycobacterium Tuberculosis Histidine Kinase Lacking an ATP-Lid Motif.
J.Biol.Chem., 288, 2013
|
|
7C7N
 
 | |
4CE1
 
 | | Hsp90 N-terminal domain bound to macrolactam analogues of radicicol. | | 分子名称: | 15-Chloro-16,18-dihydroxy-2-methyl-3,4,7,8,9,10,11,12-octahydrobenz[c][1]azacyclohexadecine-1,13(2H,14H)-dione, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | 著者 | Roe, S.M, Parry-Morris, S, Prodromou, C. | | 登録日 | 2013-11-08 | | 公開日 | 2014-01-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Synthesis of Macrolactam Analogues of Radicicol and Their Binding to Heat Shock Protein Hsp90.
Org.Biomol.Chem., 12, 2014
|
|
4CE2
 
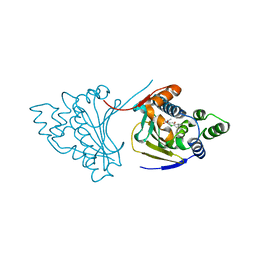 | | Hsp90 N-terminal domain bound to macrolactam analogues of radicicol. | | 分子名称: | (9E)-19-CHLORANYL-13-METHYL-16,18-BIS(OXIDANYL)-13-AZABICYCLO[13.4.0]NONADECA-1(15),9,16,18-TETRAENE-3,14-DIONE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | 著者 | Roe, S.M, Parry-Morris, S, Prodromou, C. | | 登録日 | 2013-11-08 | | 公開日 | 2014-01-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Synthesis of macrolactam analogues of radicicol and their binding to heat shock protein Hsp90.
Org. Biomol. Chem., 12, 2014
|
|
4HYP
 
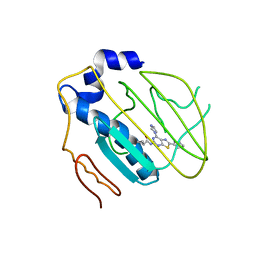 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. | | 分子名称: | DNA gyrase subunit B, MAGNESIUM ION, N-[7-(1H-imidazol-1-yl)-2-(pyridin-3-yl)[1,3]thiazolo[5,4-d]pyrimidin-5-yl]cyclopropanecarboxamide | | 著者 | Bensen, D.C, Creighton, C.J, Tari, L.W. | | 登録日 | 2012-11-13 | | 公開日 | 2013-02-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4HZ5
 
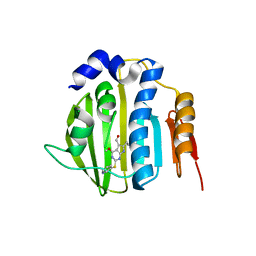 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity | | 分子名称: | 6-ethyl-4-methoxy-2-(pyridin-3-ylsulfanyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbaldehyde, DNA topoisomerase IV, B subunit | | 著者 | Bensen, D.C, Creighton, C.J, Tari, L.W. | | 登録日 | 2012-11-14 | | 公開日 | 2013-02-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4K4O
 
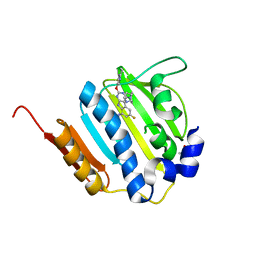 | | The DNA Gyrase B ATP binding domain of Enterococcus faecalis in complex with a small molecule inhibitor | | 分子名称: | 6-fluoro-4-[(3aR,6aR)-hexahydropyrrolo[3,4-b]pyrrol-5(1H)-yl]-N-methyl-2-[(2-methylpyrimidin-5-yl)oxy]-9H-pyrimido[4,5-b]indol-8-amine, DNA gyrase subunit B, TERTIARY-BUTYL ALCOHOL | | 著者 | Bensen, D.C, Akers-Rodriguez, S, Lam, T, Tari, L.W. | | 登録日 | 2013-04-12 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | A new class of type IIA topoisomerase inhibitors with broad-spectrum antibacterial
activity
To be Published
|
|
4KSG
 
 | | Dna gyrase atp binding domain of enterococcus faecalis in complex with a small molecule inhibitor (4-[(1S,5R,6R)-6-AMINO-1-METHYL-3-AZABICYCLO[3.2.0]HEPT-3-YL]-6-FLUORO-N-METHYL-2-[(2-METHYLPYRIMIDIN-5-YL)OXY]-9H-PYRIMIDO[4,5-B]INDOL-8-AMINE) | | 分子名称: | 4-[(1S,5R,6R)-6-amino-1-methyl-3-azabicyclo[3.2.0]hept-3-yl]-6-fluoro-N-methyl-2-[(2-methylpyrimidin-5-yl)oxy]-9H-pyrimido[4,5-b]indol-8-amine, DNA gyrase subunit B, TERTIARY-BUTYL ALCOHOL | | 著者 | Bensen, D.C, Akers-Rodriguez, S, Tari, L.W. | | 登録日 | 2013-05-17 | | 公開日 | 2014-01-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | A new class of type iia topoisomerase inhibitors with
broad-spectrum antibacterial activity
To be Published
|
|
4HXW
 
 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. | | 分子名称: | (3R)-1-[5-chloro-6-ethyl-2-(pyrido[2,3-b]pyrazin-7-ylsulfanyl)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]pyrrolidin-3-amine, DNA gyrase subunit B, TERTIARY-BUTYL ALCOHOL | | 著者 | Bensen, D.C, Trzoss, M, Tari, L.W. | | 登録日 | 2012-11-12 | | 公開日 | 2013-02-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
