8SQK
 
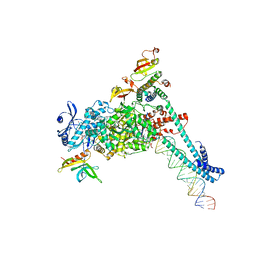 | | SARS-CoV-2 replication-transcription complex bound to RNA-nsp9 and GDP-betaS, as a pre-catalytic deRNAylation/mRNA capping intermediate | | 分子名称: | 5'-O-[(R)-hydroxy(thiophosphonooxy)phosphoryl]guanosine, MAGNESIUM ION, Non-structural protein 7, ... | | 著者 | Small, G.I, Darst, S.A, Campbell, E.A. | | 登録日 | 2023-05-04 | | 公開日 | 2023-11-22 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | Structural and functional insights into the enzymatic plasticity of the SARS-CoV-2 NiRAN domain.
Mol.Cell, 83, 2023
|
|
3NMD
 
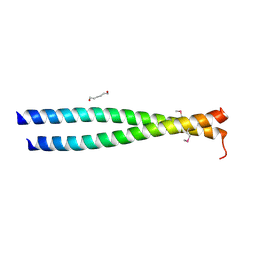 | | Crystal structure of the leucine zipper domain of cGMP dependent protein kinase I beta | | 分子名称: | GLYCEROL, HEXANE-1,6-DIOL, cGMP Dependent PRotein Kinase | | 著者 | Kim, C, Casteel, D.E, Smith-Nguyen, E.V, Sankaran, B, Berkeley Structural Genomics Center (BSGC) | | 登録日 | 2010-06-22 | | 公開日 | 2010-09-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.272 Å) | | 主引用文献 | A crystal structure of the cyclic GMP-dependent protein kinase I{beta} dimerization/docking domain reveals molecular details of isoform-specific anchoring.
J.Biol.Chem., 285, 2010
|
|
1B88
 
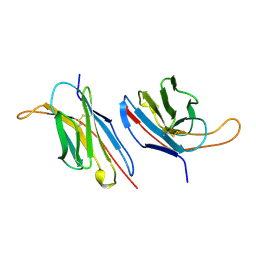 | | V-ALPHA 2.6 MOUSE T CELL RECEPTOR (TCR) DOMAIN | | 分子名称: | T CELL RECEPTOR V-ALPHA DOMAIN | | 著者 | Plaksin, D, Chacko, S, Navaza, J, Margulies, D.H, Padlan, E.A. | | 登録日 | 1999-02-09 | | 公開日 | 1999-02-16 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The X-ray crystal structure of a Valpha2.6Jalpha38 mouse T cell receptor domain at 2.5 A resolution: alternate modes of dimerization and crystal packing.
J.Mol.Biol., 289, 1999
|
|
4WKF
 
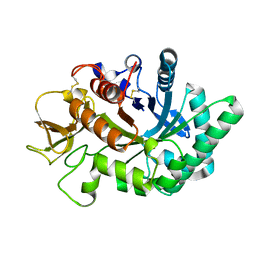 | | Crystal structure of human chitotriosidase-1 catalytic domain in complex with chitobiose (2.5mM) at 1.10 A resolution | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitotriosidase-1 | | 著者 | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | 登録日 | 2014-10-02 | | 公開日 | 2015-07-08 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.101 Å) | | 主引用文献 | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6S0X
 
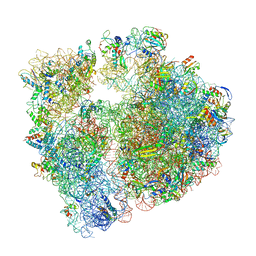 | | Erythromycin Resistant Staphylococcus aureus 70S ribosome (delta R88 A89 uL22) in complex with erythromycin. | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Halfon, Y, Matozv, D, Eyal, Z, Bashan, A, Zimmerman, E, Kjeldgaard, J, Ingmer, H, Yonath, A. | | 登録日 | 2019-06-18 | | 公開日 | 2019-08-21 | | 実験手法 | ELECTRON MICROSCOPY (2.425 Å) | | 主引用文献 | Exit tunnel modulation as resistance mechanism of S. aureus erythromycin resistant mutant.
Sci Rep, 9, 2019
|
|
6RXX
 
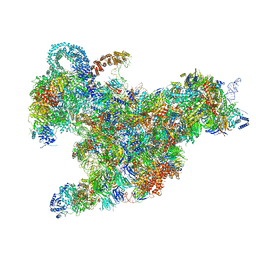 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state C, Poly-Ala | | 分子名称: | 35S ribosomal RNA, 40S ribosomal protein S1, 40S ribosomal protein S11-like protein, ... | | 著者 | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | 登録日 | 2019-06-10 | | 公開日 | 2019-08-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (7.1 Å) | | 主引用文献 | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
2J5D
 
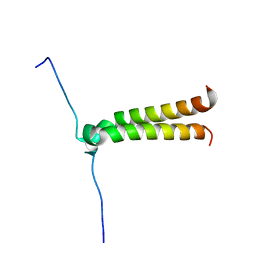 | | NMR structure of BNIP3 transmembrane domain in lipid bicelles | | 分子名称: | BCL2/ADENOVIRUS E1B 19 KDA PROTEIN-INTERACTING PROTEIN 3 | | 著者 | Bocharov, E.V, Pustovalova, Y.E, Volynsky, P.E, Maslennikov, I.V, Goncharuk, M.V, Ermolyuk, Y.S, Arseniev, A.S. | | 登録日 | 2006-09-14 | | 公開日 | 2007-04-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Unique dimeric structure of BNip3 transmembrane domain suggests membrane permeabilization as a cell death trigger.
J. Biol. Chem., 282, 2007
|
|
6S5A
 
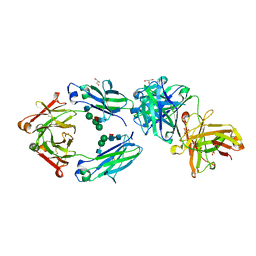 | | CRYSTAL STRUCTURE OF FC P329G LALA WITH ANTI FC P329G FAB | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Fc P329G LALA, ... | | 著者 | Ehler, A, Darowski, D, Jost, C, Stubenrauch, K, Wessels, U, Benz, J, Birk, M, Freimoser-Grundschober, A, Bruenker, P, Moessner, E, Umana, P, Kobold, S, Klein, C. | | 登録日 | 2019-07-01 | | 公開日 | 2019-09-25 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | P329G-CAR-J: a novel Jurkat-NFAT-based CAR-T reporter system recognizing the P329G Fc mutation.
Protein Eng.Des.Sel., 32, 2019
|
|
6S5V
 
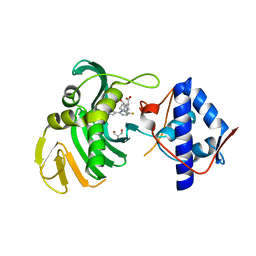 | | Crystal structure of the Cap-Midlink region of the H5N1 Influenza A virus polymerase in complex with a Cap-domain binding analogue | | 分子名称: | (1~{S},2~{S},3~{S},6~{R})-2-[[2-[5,7-bis(fluoranyl)-1~{H}-indol-3-yl]-5-fluoranyl-pyrimidin-4-yl]amino]-3,6-dimethyl-cyclohexane-1-carboxylic acid, GLYCEROL, POTASSIUM ION, ... | | 著者 | Keown, J.R, Fodor, E, Grimes, J.M. | | 登録日 | 2019-07-02 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Design, Synthesis, and Biological Evaluation of Novel Indoles Targeting the Influenza PB2 Cap Binding Region.
J.Med.Chem., 62, 2019
|
|
8HQR
 
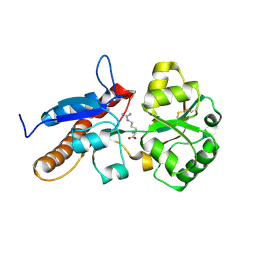 | |
5A5K
 
 | | AtGSTF2 from Arabidopsis thaliana in complex with camalexin | | 分子名称: | (2Z)-2-indol-3-ylidene-3H-1,3-thiazole, GLUTATHIONE S-TRANSFERASE F2 | | 著者 | Ahmad, L, Rylott, E, Bruce, N.C, Edwards, R, Grogan, G. | | 登録日 | 2015-06-18 | | 公開日 | 2016-06-29 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.77 Å) | | 主引用文献 | Structural evidence for Arabidopsis glutathione transferase AtGSTF2 functioning as a transporter of small organic ligands.
FEBS Open Bio, 7, 2017
|
|
5A5V
 
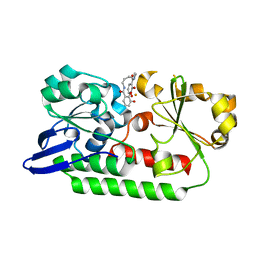 | | A complex of the synthetic siderophore analogue Fe(III)-6-LICAM with the CeuE periplasmic protein from Campylobacter jejuni | | 分子名称: | ENTEROCHELIN UPTAKE PERIPLASMIC BINDING PROTEIN, FE (III) ION, N,N'-hexane-1,4-diylbis(2,3-dihydroxybenzamide) | | 著者 | Blagova, E, Hughes, A, Moroz, O.V, Raines, D.J, Wilde, E.J, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | 登録日 | 2015-06-22 | | 公開日 | 2016-07-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
5KNK
 
 | | Lipid A secondary acyltransferase LpxM from Acinetobacter baumannii with catalytic residue substitution (E127A) | | 分子名称: | DODECYL-BETA-D-MALTOSIDE, GLYCEROL, Lipid A biosynthesis lauroyl acyltransferase, ... | | 著者 | Dovala, D.L, Hu, Q, Metzger IV, L.E. | | 登録日 | 2016-06-28 | | 公開日 | 2016-09-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-guided enzymology of the lipid A acyltransferase LpxM reveals a dual activity mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ZR1
 
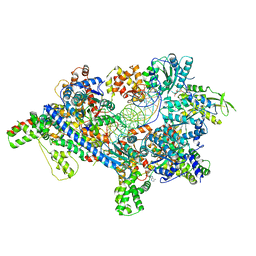 | | Saccharomyces Cerevisiae Origin Recognition Complex Bound to a 72-bp Origin DNA containing ACS and B1 element | | 分子名称: | 72bp-oring DNA, ACS305, A-rich, ... | | 著者 | Li, N, Lam, W.H, Zhai, Y, Cheng, J, Cheng, E, Zhao, Y, Gao, N, Tye, B.K. | | 登録日 | 2018-04-21 | | 公開日 | 2018-07-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure of the origin recognition complex bound to DNA replication origin.
Nature, 559, 2018
|
|
3HHA
 
 | | Crystal structure of cathepsin L in complex with AZ12878478 | | 分子名称: | ACETATE ION, Cathepsin L1, GLYCEROL, ... | | 著者 | Asaad, N, Bethel, P.A, Coulson, M.D, Dawson, J, Ford, S.J, Gerhardt, S, Grist, M, Hamlin, G.A, James, M.J, Jones, E.V, Karoutchi, G.I, Kenny, P.W, Morley, A.D, Oldham, K, Rankine, N, Ryan, D, Wells, S.L, Wood, L, Augustin, M, Krapp, S, Simader, H, Steinbacher, S. | | 登録日 | 2009-05-15 | | 公開日 | 2009-06-23 | | 最終更新日 | 2021-10-13 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | Dipeptidyl nitrile inhibitors of Cathepsin L.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3HO0
 
 | | Crystal structure of the PPARgamma-LBD complexed with a new aryloxy-3phenylpropanoic acid | | 分子名称: | (2S)-2-(4-phenethylphenoxy)-3-phenyl-propanoic acid, Peroxisome proliferator-activated receptor gamma | | 著者 | Pochetti, G, Montanari, R, Mazza, F, Loiodice, F, Fracchiolla, G, Laghezza, A, Lavecchia, A, Novellino, E. | | 登録日 | 2009-06-01 | | 公開日 | 2009-10-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | New 2-Aryloxy-3-phenyl-propanoic Acids As Peroxisome Proliferator-Activated Receptors alpha/gamma Dual Agonists with Improved Potency and Reduced Adverse Effects on Skeletal Muscle Function
J.Med.Chem., 52, 2009
|
|
6MBP
 
 | | GLP Methyltransferase with Inhibitor EML741- P3121 Crystal Form | | 分子名称: | 1,2-ETHANEDIOL, 2-cyclohexyl-7-methoxy-N-[1-(propan-2-yl)piperidin-4-yl]-8-[3-(pyrrolidin-1-yl)propoxy]-3H-1,4-benzodiazepin-5-amine, Histone-lysine N-methyltransferase EHMT1, ... | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2018-08-30 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.947 Å) | | 主引用文献 | Discovery of a Novel Chemotype of Histone Lysine Methyltransferase EHMT1/2 (GLP/G9a) Inhibitors: Rational Design, Synthesis, Biological Evaluation, and Co-crystal Structure.
J. Med. Chem., 62, 2019
|
|
5L5Q
 
 | |
1B8M
 
 | | BRAIN DERIVED NEUROTROPHIC FACTOR, NEUROTROPHIN-4 | | 分子名称: | PROTEIN (BRAIN DERIVED NEUROTROPHIC FACTOR), PROTEIN (NEUROTROPHIN-4) | | 著者 | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y, Choe, S. | | 登録日 | 1999-02-01 | | 公開日 | 1999-02-09 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | The structures of the neurotrophin 4 homodimer and the brain-derived neurotrophic factor/neurotrophin 4 heterodimer reveal a common Trk-binding site.
Protein Sci., 8, 1999
|
|
5L66
 
 | |
6RS0
 
 | | GOLGI ALPHA-MANNOSIDASE II in complex with (2S,3S,4R,5R)-1-(2-(Benzyloxy)ethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol | | 分子名称: | (2~{S},3~{S},4~{R})-2-(hydroxymethyl)-1-(2-phenylmethoxyethyl)piperidine-3,4,5-triol, 1,2-ETHANEDIOL, Alpha-mannosidase 2, ... | | 著者 | Armstrong, Z, Lahav, D, Johnson, R, Kuo, C.L, Beenakker, T.J.M, de Boer, C, Wong, C.S, van Rijssel, E.R, Debets, M, Geurink, P.P, Ovaa, H, van der Stelt, M, Codee, J.D.C, Aerts, J.M.F.G, Wu, L, Overkleeft, H.S, Davies, G.J. | | 登録日 | 2019-05-20 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Manno- epi -cyclophellitols Enable Activity-Based Protein Profiling of Human alpha-Mannosidases and Discovery of New Golgi Mannosidase II Inhibitors.
J.Am.Chem.Soc., 142, 2020
|
|
5KRT
 
 | |
6RMX
 
 | | Human Carbonic anhydrase II mutant T199V bound by 2-[(1S)-2,3-Dihydro-1H-inden-1-ylamino]-3,5,6-trifluoro-4-[(2-hydroxyethyl)thio]benzenesulfonamide | | 分子名称: | 2-[(1S)-2,3-dihydro-1H-inden-1-ylamino]-3,5,6-trifluoro-4-[(2-hydroxyethyl)sulfanyl]benzenesulfonamide, BENZOIC ACID, Carbonic anhydrase 2, ... | | 著者 | Smirnov, A, Paketuryte, V, Manakova, E, Grazulis, S. | | 登録日 | 2019-05-07 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Human Carbonic anhydrase II mutant T199V bound by 2-[(1S)-2,3-Dihydro-1H-inden-1-ylamino]-3,5,6-trifluoro-4-[(2-hydroxyethyl)thio]benzenesulfonamide
To Be Published
|
|
8HYG
 
 | |
1BBJ
 
 | |
