5DOR
 
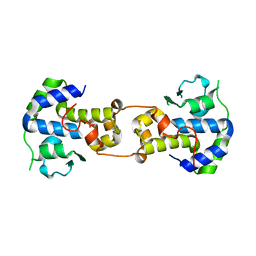 | | P2 Integrase catalytic domain in space group P21 | | 分子名称: | Integrase, PHOSPHATE ION, ZINC ION | | 著者 | Skaar, K, Claesson, M, Odegrip, R, Haggard-Ljungquist, E, Hogbom, M. | | 登録日 | 2015-09-11 | | 公開日 | 2015-12-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the bacteriophage P2 integrase catalytic domain.
Febs Lett., 589, 2015
|
|
4U0V
 
 | |
4U0W
 
 | |
3KQL
 
 | |
3KQN
 
 | |
3KQU
 
 | |
5E0U
 
 | |
8RU2
 
 | | Structure of the F-actin barbed end bound by formin mDia1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | 著者 | Oosterheert, W, Boiero Sanders, M, Funk, J, Prumbaum, D, Raunser, S, Bieling, P. | | 登録日 | 2024-01-29 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | Molecular mechanism of actin filament elongation by formins.
Science, 384, 2024
|
|
8RTY
 
 | | Structure of the F-actin barbed end bound by Cdc12 and profilin (ring complex) at a resolution of 6.3 Angstrom | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | 著者 | Oosterheert, W, Boiero Sanders, M, Funk, J, Prumbaum, D, Raunser, S, Bieling, P. | | 登録日 | 2024-01-29 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (6.25 Å) | | 主引用文献 | Molecular mechanism of actin filament elongation by formins.
Science, 384, 2024
|
|
5E0V
 
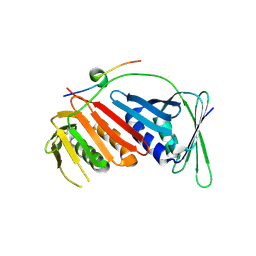 | |
8HIO
 
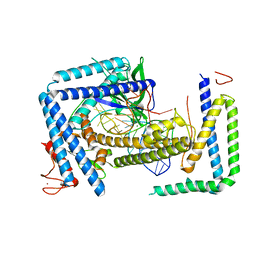 | | Cryo-EM structure of the Cas12m2-crRNA binary complex | | 分子名称: | Cas12m2, MAGNESIUM ION, RNA (56-MER), ... | | 著者 | Omura, N.S, Nakagawa, R, Wu, Y.W, Sudfeld, C, Warren, V.R, Hirano, H, Kusakizako, T, Kise, Y, Lebbink, H.G.J, Itoh, Y, Oost, V.D.J, Nureki, O. | | 登録日 | 2022-11-21 | | 公開日 | 2023-04-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (3.73 Å) | | 主引用文献 | Mechanistic and evolutionary insights into a type V-M CRISPR-Cas effector enzyme.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3KP3
 
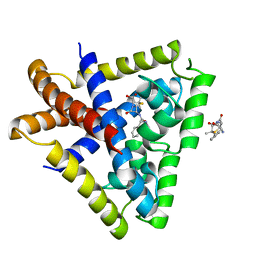 | | Staphylococcus epidermidis in complex with ampicillin | | 分子名称: | (2S,5R,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, Transcriptional regulator TcaR | | 著者 | Chang, Y.M, Chen, C.K, Wang, A.H. | | 登録日 | 2009-11-15 | | 公開日 | 2010-06-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural study of TcaR and its complexes with multiple antibiotics from Staphylococcus epidermidis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4W97
 
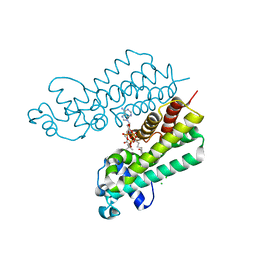 | | Structure of ketosteroid transcriptional regulator KstR2 of Mycobacterium tuberculosis | | 分子名称: | CHLORIDE ION, HTH-type transcriptional repressor KstR2, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] 3-[(3aS,4S,7aS)-7a-methyl-1,5-bis(oxidanylidene)-2,3,3a,4,6,7-hexahydroinden-4-yl]propanethioate | | 著者 | Stogios, P.J, Evdokimova, E, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-08-27 | | 公開日 | 2014-11-26 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural and Functional Characterization of a Ketosteroid Transcriptional Regulator of Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
1VZK
 
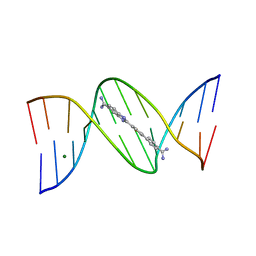 | | A Thiophene Based Diamidine Forms a "Super" AT Binding Minor Groove Agent | | 分子名称: | 2-(5-{4-[AMINO(IMINO)METHYL]PHENYL}-2-THIENYL)-1H-BENZIMIDAZOLE-6- CARBOXIMIDAMIDE DIHYDROCHLORIDE, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP *CP*G)-3', MAGNESIUM ION | | 著者 | Mallena, S, Lee, M.P.H, Bailly, C, Neidle, S, Kumar, A, Boykin, D.W, Wilson, W.D. | | 登録日 | 2004-05-20 | | 公開日 | 2004-10-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Thiophene-Based Diamidine Forms a "Super" at Binding Minor Groove Agent
J.Am.Chem.Soc., 142, 2004
|
|
8BUM
 
 | | Structure of DDB1 bound to DS15-engaged CDK12-cyclin K | | 分子名称: | (2R)-2-[[6-(5-naphthalen-1-ylpentylamino)-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-20 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.36 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU7
 
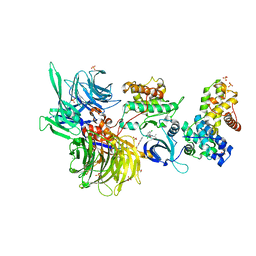 | | Structure of DDB1 bound to 21195-engaged CDK12-cyclin K | | 分子名称: | 1,2-ETHANEDIOL, 1-[2,6-bis(chloranyl)phenyl]-6-[[4-(2-hydroxyethyloxy)phenyl]methyl]-3-propan-2-yl-5H-pyrazolo[3,4-d]pyrimidin-4-one, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.245 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUN
 
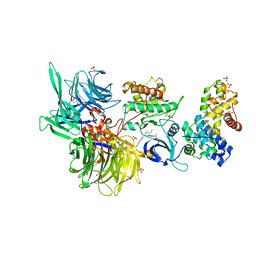 | | Structure of DDB1 bound to DS16-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[6-[(4-phenylphenyl)methylamino]-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.08 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU2
 
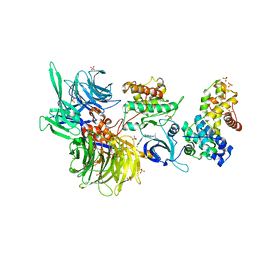 | | Structure of DDB1 bound to DS18-engaged CDK12-cyclin K | | 分子名称: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.13 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUA
 
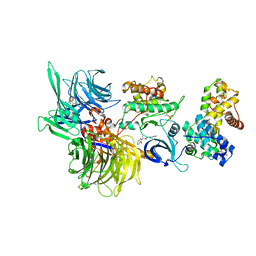 | | Structure of DDB1 bound to 919278-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-~{N}-(1~{H}-benzimidazol-2-yl)-2-(3-oxidanylidene-1~{H}-isoindol-2-yl)propanamide, CITRIC ACID, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.193 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUI
 
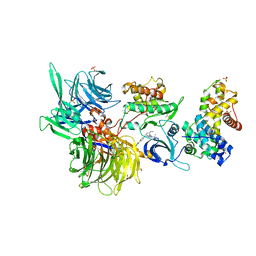 | | Structure of DDB1 bound to DRF-053-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[9-propan-2-yl-6-[(4-pyridin-2-ylphenyl)amino]purin-2-yl]amino]butan-1-ol, 1,2-ETHANEDIOL, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUR
 
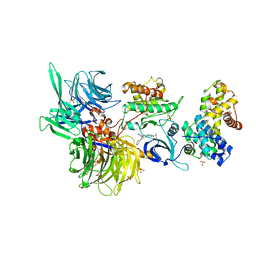 | | Structure of DDB1 bound to DS50-engaged CDK12-cyclin K | | 分子名称: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.64 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUF
 
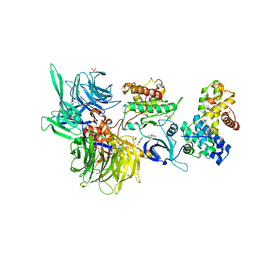 | | Structure of DDB1 bound to Z12-engaged CDK12-cyclin K | | 分子名称: | 2-(6,7-dihydro-4~{H}-thieno[3,2-c]pyridin-5-ylmethyl)-6,7-dimethoxy-3~{H}-quinazolin-4-one, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU1
 
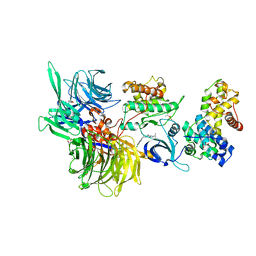 | | Structure of DDB1 bound to DS17-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[6-[[5,6-bis(chloranyl)-1~{H}-benzimidazol-2-yl]methylamino]-9-(1-methylpyrazol-4-yl)purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
6L87
 
 | |
6DK4
 
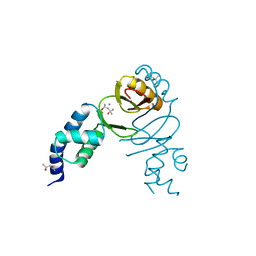 | | Crystal structure of Campylobacter jejuni peroxide stress regulator | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Ferric uptake regulation protein, MANGANESE (II) ION, ... | | 著者 | Sarvan, S, Brunzelle, J.S, Couture, J.F. | | 登録日 | 2018-05-28 | | 公開日 | 2018-06-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Crystal structure of Campylobacter jejuni peroxide regulator.
FEBS Lett., 592, 2018
|
|
