1TKG
 
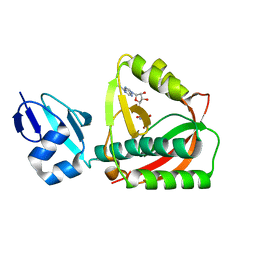 | | Crystal structure of the editing domain of threonyl-tRNA synthetase complexed with an analog of seryladenylate | | 分子名称: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, Threonyl-tRNA synthetase | | 著者 | Dock-Bregeon, A.C, Rees, B, Torres-Larios, A, Bey, G, Caillet, J, Moras, D. | | 登録日 | 2004-06-08 | | 公開日 | 2004-11-30 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Achieving Error-Free Translation; The Mechanism of Proofreading of Threonyl-tRNA Synthetase at Atomic Resolution.
Mol.Cell, 16, 2004
|
|
7SYN
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head opening. Structure 8(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, HCV IRES, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYJ
 
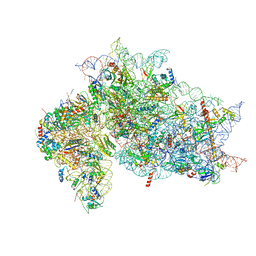 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 4(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYL
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, closed conformation. Structure 6(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYX
 
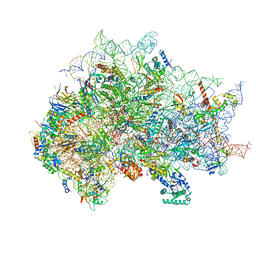 | | Structure of the delta dII IRES eIF5B-containing 48S initiation complex, closed conformation. Structure 15(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S24, 40S ribosomal protein S25, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-03-08 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYW
 
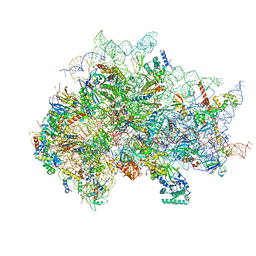 | | Structure of the wt IRES eIF5B-containing 48S initiation complex, closed conformation. Structure 15(wt) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-02-01 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYG
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 1(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYI
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 3(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYM
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head opening. Structure 7(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Comprehensive structural overview of the HCV IRES-mediated translation initiation pathway
To Be Published
|
|
7ZUB
 
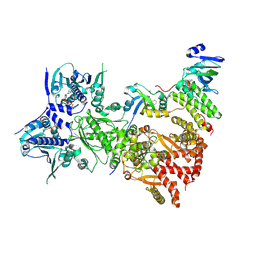 | | Cryo-EM structure of the indirubin-bound Hsp90-XAP2-AHR complex | | 分子名称: | (3~{Z})-3-(3-oxidanylidene-1~{H}-indol-2-ylidene)-1~{H}-indol-2-one, ADENOSINE-5'-DIPHOSPHATE, AH receptor-interacting protein, ... | | 著者 | Gruszczyk, J, Savva, C.G, Lai-Kee-Him, J, Bous, J, Ancelin, A, Kwong, H.S, Grandvuillemin, L, Bourguet, W. | | 登録日 | 2022-05-12 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Cryo-EM structure of the agonist-bound Hsp90-XAP2-AHR cytosolic complex.
Nat Commun, 13, 2022
|
|
7T3A
 
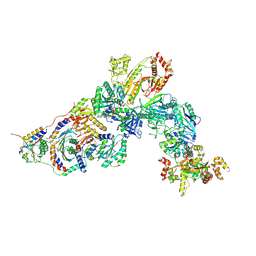 | | GATOR1-RAG-RAGULATOR - Inhibitory Complex | | 分子名称: | GATOR complex protein DEPDC5, GATOR complex protein NPRL2, GATOR complex protein NPRL3, ... | | 著者 | Egri, S.B, Shen, K. | | 登録日 | 2021-12-07 | | 公開日 | 2022-04-06 | | 最終更新日 | 2022-06-01 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Cryo-EM structures of the human GATOR1-Rag-Ragulator complex reveal a spatial-constraint regulated GAP mechanism.
Mol.Cell, 82, 2022
|
|
7T3B
 
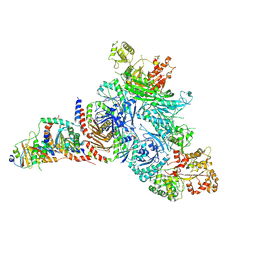 | | GATOR1-RAG-RAGULATOR - GAP Complex | | 分子名称: | ALUMINUM FLUORIDE, GATOR complex protein DEPDC5, GATOR complex protein NPRL2, ... | | 著者 | Egri, S.B, Shen, K. | | 登録日 | 2021-12-07 | | 公開日 | 2022-04-06 | | 最終更新日 | 2022-06-01 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM structures of the human GATOR1-Rag-Ragulator complex reveal a spatial-constraint regulated GAP mechanism.
Mol.Cell, 82, 2022
|
|
7T3C
 
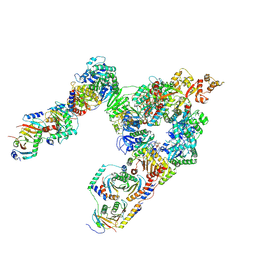 | | GATOR1-RAG-RAGULATOR - Dual Complex | | 分子名称: | ALUMINUM FLUORIDE, GATOR complex protein DEPDC5, GATOR complex protein NPRL2, ... | | 著者 | Egri, S.B, Shen, K. | | 登録日 | 2021-12-07 | | 公開日 | 2022-04-06 | | 最終更新日 | 2022-06-01 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Cryo-EM structures of the human GATOR1-Rag-Ragulator complex reveal a spatial-constraint regulated GAP mechanism.
Mol.Cell, 82, 2022
|
|
5LKS
 
 | | Structure-function insights reveal the human ribosome as a cancer target for antibiotics | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | 著者 | Myasnikov, A.G, Natchiar, S.K, Nebout, M, Hazemann, I, Imbert, V, Khatter, H, Peyron, J.-F, Klaholz, B.P. | | 登録日 | 2016-07-23 | | 公開日 | 2017-04-26 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure-function insights reveal the human ribosome as a cancer target for antibiotics.
Nat Commun, 7, 2016
|
|
8VEU
 
 | |
8VET
 
 | |
8VEX
 
 | |
8VEY
 
 | |
8VEW
 
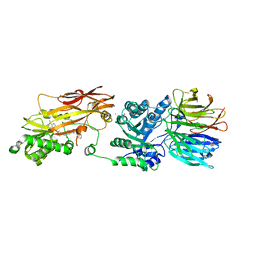 | | Crystal structure of PRMT5:MEP50 in complex with MTA and oxamide compound 24 | | 分子名称: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, 5-{2-[(2R,5S)-5-methyl-2-phenylpiperidin-1-yl](oxo)acetamido}pyridine-3-carboxamide, CHLORIDE ION, ... | | 著者 | Whittington, D.A. | | 登録日 | 2023-12-20 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Discovery of TNG908: A Selective, Brain Penetrant, MTA-Cooperative PRMT5 Inhibitor That Is Synthetically Lethal with MTAP -Deleted Cancers.
J.Med.Chem., 67, 2024
|
|
8VEO
 
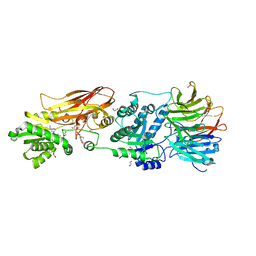 | | Crystal structure of PRMT5:MEP50 in complex with MTA | | 分子名称: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Whittington, D.A. | | 登録日 | 2023-12-20 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Discovery of TNG908: A Selective, Brain Penetrant, MTA-Cooperative PRMT5 Inhibitor That Is Synthetically Lethal with MTAP -Deleted Cancers.
J.Med.Chem., 67, 2024
|
|
9BIV
 
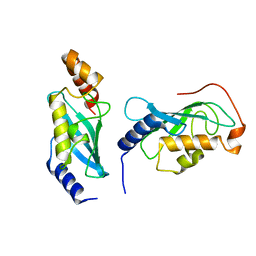 | |
7ZR5
 
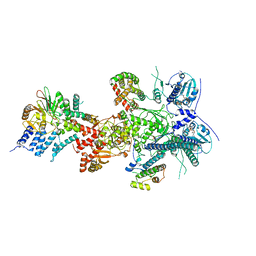 | |
7ZR6
 
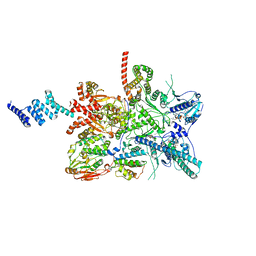 | |
5MQF
 
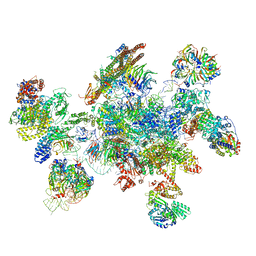 | | Cryo-EM structure of a human spliceosome activated for step 2 of splicing (C* complex) | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ATP-dependent RNA helicase DHX8, Cell division cycle 5-like protein, ... | | 著者 | Bertram, K, Hartmuth, K, Kastner, B. | | 登録日 | 2016-12-20 | | 公開日 | 2017-03-22 | | 最終更新日 | 2018-11-21 | | 実験手法 | ELECTRON MICROSCOPY (5.9 Å) | | 主引用文献 | Cryo-EM structure of a human spliceosome activated for step 2 of splicing.
Nature, 542, 2017
|
|
5V3B
 
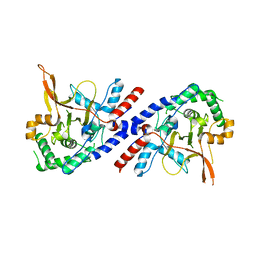 | |
