4AUU
 
 | | Crystal structure of apo FimH lectin domain at 1.5 A resolution | | 分子名称: | 1,2-ETHANEDIOL, FIMH, NICKEL (II) ION, ... | | 著者 | Wellens, A, Lahmann, M, Touaibia, M, Vaucher, J, Oscarson, S, Roy, R, Remaut, H, Bouckaert, J. | | 登録日 | 2012-05-22 | | 公開日 | 2012-06-27 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The Tyrosine Gate as a Potential Entropic Lever in the Receptor-Binding Site of the Bacterial Adhesin Fimh.
Biochemistry, 51, 2012
|
|
1M3G
 
 | |
6MCE
 
 | |
3EPB
 
 | | Human AdoMetDC E256Q mutant complexed with putrescine | | 分子名称: | 1,4-DIAMINOBUTANE, PYRUVIC ACID, S-adenosylmethionine decarboxylase alpha chain, ... | | 著者 | Bale, S, Lopez, M.M, Makhatadze, G.I, Fang, Q, Pegg, A.E, Ealick, S.E. | | 登録日 | 2008-09-29 | | 公開日 | 2008-12-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural Basis for Putrescine Activation of Human S-Adenosylmethionine Decarboxylase.
Biochemistry, 47, 2008
|
|
2JBL
 
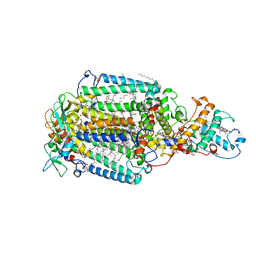 | | PHOTOSYNTHETIC REACTION CENTER FROM BLASTOCHLORIS VIRIDIS | | 分子名称: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | 著者 | Lancaster, C.R.D. | | 登録日 | 2006-12-08 | | 公開日 | 2007-03-13 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A Comparison of Stigmatellin Conformations, Free and Bound to the Photosynthetic Reaction Center and the Cytochrome Bc(1) Complex.
J.Mol.Biol., 368, 2007
|
|
1KV2
 
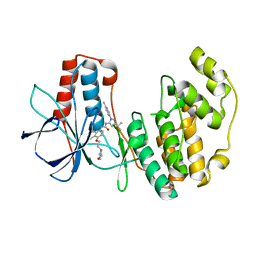 | | Human p38 MAP Kinase in Complex with BIRB 796 | | 分子名称: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, p38 MAP kinase | | 著者 | Pargellis, C, Tong, L, Churchill, L, Cirillo, P.F, Gilmore, T, Graham, A.G, Grob, P.M, Hickey, E.R, Moss, N, Pav, S, Regan, J. | | 登録日 | 2002-01-23 | | 公開日 | 2002-03-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site.
Nat.Struct.Biol., 9, 2002
|
|
7K5E
 
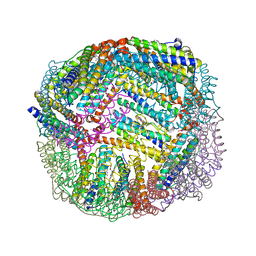 | | 1.75 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor JAG-5-7 | | 分子名称: | 4-{[(5-fluoro-2-hydroxyphenyl)methyl]amino}-1H-isoindole-1,3(2H)-dione, Ferroxidase, POTASSIUM ION, ... | | 著者 | Lovell, S, Battaile, K.P, Soldano, A, Punchi-Hewage, A, Meraz, K, Annor-Gyamfi, J.K, Yao, H, Bunce, R.A, Rivera, M. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Small Molecule Inhibitors of the Bacterioferritin (BfrB)-Ferredoxin (Bfd) Complex Kill Biofilm-Embedded Pseudomonas aeruginosa Cells.
Acs Infect Dis., 7, 2021
|
|
7K5F
 
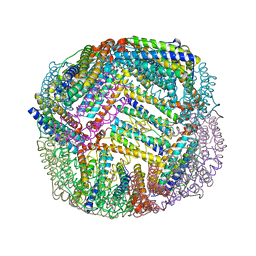 | | 1.90 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor KM-5-50 | | 分子名称: | 4-{[(3-chloro-5-hydroxyphenyl)methyl]amino}-1H-isoindole-1,3(2H)-dione, Ferroxidase, POTASSIUM ION, ... | | 著者 | Lovell, S, Battaile, K.P, Soldano, A, Punchi-Hewage, A, Meraz, K, Annor-Gyamfi, J.K, Yao, H, Bunce, R.A, Rivera, M. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Small Molecule Inhibitors of the Bacterioferritin (BfrB)-Ferredoxin (Bfd) Complex Kill Biofilm-Embedded Pseudomonas aeruginosa Cells.
Acs Infect Dis., 7, 2021
|
|
5IUS
 
 | | Crystal structure of human PD-L1 in complex with high affinity PD-1 mutant | | 分子名称: | CHLORIDE ION, Programmed cell death 1 ligand 1, Programmed cell death protein 1 | | 著者 | Pascolutti, R, Sun, X, Kao, J, Maute, R, Ring, A.M, Bowman, G.R, Kruse, A.C. | | 登録日 | 2016-03-18 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.889 Å) | | 主引用文献 | Structure and Dynamics of PD-L1 and an Ultra-High-Affinity PD-1 Receptor Mutant.
Structure, 24, 2016
|
|
2WB6
 
 | | Crystal structure of AFV1-102, a protein from the Acidianus Filamentous Virus 1 | | 分子名称: | AFV1-102, CHLORIDE ION | | 著者 | Keller, J, Leulliot, N, Collinet, B, Campanacci, V, Cambillau, C, Pranghisvilli, D, van Tilbeurgh, H. | | 登録日 | 2009-02-22 | | 公開日 | 2009-03-03 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal Structure of Afv1-102, a Protein from the Acidianus Filamentous Virus 1.
Protein Sci., 18, 2009
|
|
5EU7
 
 | | Crystal structure of HIV-1 integrase catalytic core in complex with Fab | | 分子名称: | FAB Heavy Chain, FAB light chain, Integrase | | 著者 | Galilee, M, Griner, S.L, Stroud, R.M, Alian, A. | | 登録日 | 2015-11-18 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | The Preserved HTH-Docking Cleft of HIV-1 Integrase Is Functionally Critical.
Structure, 24, 2016
|
|
4LWQ
 
 | | Crystal structure of native peptidyl t-RNA hydrolase from Acinetobacter baumannii at 1.38A resolution | | 分子名称: | GLYCEROL, Peptidyl-tRNA hydrolase | | 著者 | Kaushik, S, Singh, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | 登録日 | 2013-07-28 | | 公開日 | 2013-08-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Crystal structure of native peptidyl t-RNA hydrolase from Acinetobacter baumannii at 1.38A resolution
To be Published
|
|
6TL9
 
 | | CRYSTAL STRUCTURE OF LECTIN-LIKE OX-LDL RECEPTOR 1 IN COMPLEX WITH BI-0115 | | 分子名称: | 9-chloranyl-5-propyl-11~{H}-pyrido[2,3-b][1,4]benzodiazepin-6-one, GLYCEROL, Oxidized low-density lipoprotein receptor 1 | | 著者 | Nar, H, Fiegen, D, Schnapp, G. | | 登録日 | 2019-12-02 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.734 Å) | | 主引用文献 | A small-molecule inhibitor of lectin-like oxidized LDL receptor-1 acts by stabilizing an inactive receptor tetramer state
Commun Chem, 2020
|
|
6AU2
 
 | | Crystal structure of SETDB1 Tudor domain with aryl triazole fragments | | 分子名称: | 1,2-ETHANEDIOL, 1-methyl-4H,6H-[1,2,4]triazolo[4,3-a][4,1]benzoxazepine, BETA-MERCAPTOETHANOL, ... | | 著者 | MADER, P, Mendoza-Sanchez, R, IQBAL, A, DONG, A, DOBROVETSKY, E, CORLESS, V.B, LIEW, S.K, TEMPEL, W, SMIL, D, DELA SENA, C.C, KENNEDY, S, DIAZ, D, HOLOWNIA, A, VEDADI, M, BROWN, P.J, SANTHAKUMAR, V, Bountra, C, Edwards, A.M, YUDIN, A.K, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | 登録日 | 2017-08-30 | | 公開日 | 2017-10-11 | | 最終更新日 | 2025-04-02 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Identification and characterization of the first fragment hits for SETDB1 Tudor domain.
Bioorg.Med.Chem., 27, 2019
|
|
1SZK
 
 | | The structure of gamma-aminobutyrate aminotransferase mutant: E211S | | 分子名称: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | 著者 | Liu, W, Peterson, P.E, Langston, J.A, Jin, X, Fisher, A.J, Toney, M.D. | | 登録日 | 2004-04-05 | | 公開日 | 2005-03-01 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Kinetic and Crystallographic Analysis of Active Site Mutants of Escherichia coligamma-Aminobutyrate Aminotransferase.
Biochemistry, 44, 2005
|
|
1KVU
 
 | | UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH UDP-PHENOL | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Thoden, J.B, Gulick, A.M, Holden, H.M. | | 登録日 | 1997-03-07 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Mechanistic roles of tyrosine 149 and serine 124 in UDP-galactose 4-epimerase from Escherichia coli.
Biochemistry, 36, 1997
|
|
2WBK
 
 | | Structure of the Michaelis complex of beta-mannosidase, Man2A, provides insight into the conformational itinerary of mannoside hydrolysis | | 分子名称: | 1,2-ETHANEDIOL, 2,4-dinitrophenyl 2-deoxy-2-fluoro-beta-D-mannopyranoside, BETA-MANNOSIDASE, ... | | 著者 | Offen, W.A, Zechel, D.L, Withers, S.G, Gilbert, H.J, Davies, G.J. | | 登録日 | 2009-03-02 | | 公開日 | 2009-03-17 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of the Michaelis Complex of Beta-Mannosidase, Man2A, Provides Insight Into the Conformational Itinerary of Mannoside Hydrolysis.
Cell(Cambridge,Mass.), 18, 2009
|
|
4PZH
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 2,3,5,6-tetrafluoro-4[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | 分子名称: | 1,2-ETHANEDIOL, 2,3,5,6-tetrafluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | 著者 | Smirnov, A, Manakova, E, Grazulis, S. | | 登録日 | 2014-03-31 | | 公開日 | 2015-01-28 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.06 Å) | | 主引用文献 | Discovery and characterization of novel selective inhibitors of carbonic anhydrase IX.
J.Med.Chem., 57, 2014
|
|
6FWT
 
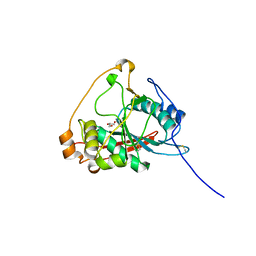 | | Crystal structure of human wild type beta-1,4-galactosyltransferase-1 (B4GalT1) in apo-open monomeric form | | 分子名称: | Beta-1,4-galactosyltransferase 1, GLYCEROL | | 著者 | Harrus, D, Kellokumpu, S, Glumoff, T. | | 登録日 | 2018-03-07 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.845 Å) | | 主引用文献 | The dimeric structure of wild-type human glycosyltransferase B4GalT1.
PLoS ONE, 13, 2018
|
|
7MIY
 
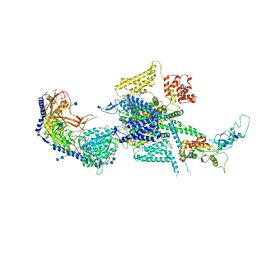 | | Human N-type voltage-gated calcium channel Cav2.2 at 3.1 Angstrom resolution | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Yan, N, Gao, S, Yao, X. | | 登録日 | 2021-04-18 | | 公開日 | 2021-07-07 | | 最終更新日 | 2025-05-28 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structure of human Ca v 2.2 channel blocked by the painkiller ziconotide.
Nature, 596, 2021
|
|
1QGM
 
 | |
1M7N
 
 | |
3XIN
 
 | |
1M8T
 
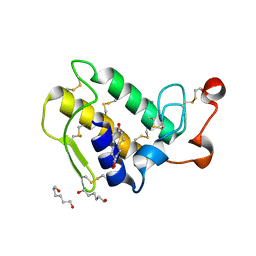 | | Structure of an acidic Phospholipase A2 from the venom of Ophiophagus hannah at 2.1 resolution from a hemihedrally twinned crystal form | | 分子名称: | CALCIUM ION, HEXANE-1,6-DIOL, Phospholipase a2 | | 著者 | Xu, S, Gu, L, Wang, Q, Shu, Y, Lin, Z. | | 登録日 | 2002-07-26 | | 公開日 | 2003-09-02 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of a king cobra phospholipase A2 determined from a hemihedrally twinned crystal.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
7FXV
 
 | | Crystal Structure of human FABP4 in complex with 6-[(4-methoxyphenyl)methyl]-2H-1,2,4-triazine-3,5-dithione | | 分子名称: | 1,2-ETHANEDIOL, 6-[(4-methoxyphenyl)methyl]-1,2,4-triazine-3,5(2H,4H)-dithione, DIMETHYL SULFOXIDE, ... | | 著者 | Ehler, A, Benz, J, Obst, U, Koch, W, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2025-08-13 | | 実験手法 | X-RAY DIFFRACTION (0.88 Å) | | 主引用文献 | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
