2C40
 
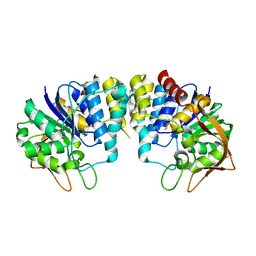 | | CRYSTAL STRUCTURE OF INOSINE-URIDINE PREFERRING NUCLEOSIDE HYDROLASE FROM BACILLUS ANTHRACIS AT 2.2A RESOLUTION | | 分子名称: | CALCIUM ION, INOSINE-URIDINE PREFERRING NUCLEOSIDE HYDROLASE FAMILY PROTEIN, alpha-D-ribofuranose | | 著者 | Moroz, O.V, Blagova, E.V, Fogg, M.J, Levdikov, V.M, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | 登録日 | 2005-10-13 | | 公開日 | 2007-02-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of Inosine-Uridine Preferring Nucleoside Hydrolase from Bacillus Anthracis at 2.2A Resolution
To be Published
|
|
5FYD
 
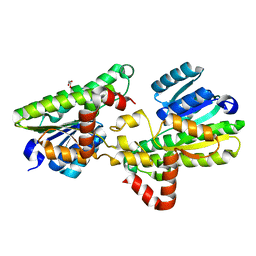 | | Structural and biochemical insights into 7beta-hydroxysteroid dehydrogenase stereoselectivity | | 分子名称: | GLYCEROL, OXIDOREDUCTASE, SHORT CHAIN DEHYDROGENASE/REDUCTASE FAMILY PROTEIN | | 著者 | Savino, S, Ferrandi, E, Forneris, F, Rovida, S, Riva, S, Monti, D, Mattevi, A. | | 登録日 | 2016-03-07 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural and Biochemical Insights Into 7Beta-Hydroxysteroid Dehydrogenase Stereoselectivity.
Proteins, 84, 2016
|
|
5GVQ
 
 | | Solution structure of the first RRM domain of human spliceosomal protein SF3b49 | | 分子名称: | Splicing factor 3B subunit 4 | | 著者 | Kuwasako, K, Nameki, N, Tsuda, K, Takahashi, M, Sato, A, Tochio, N, Inoue, M, Terada, T, Kigawa, T, Kobayashi, N, Shirouzu, M, Ito, T, Sakamoto, T, Wakamatsu, K, Guntert, P, Takahashi, S, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2016-09-06 | | 公開日 | 2017-04-12 | | 最終更新日 | 2025-03-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the first RNA recognition motif domain of human spliceosomal protein SF3b49 and its mode of interaction with a SF3b145 fragment.
Protein Sci., 26, 2017
|
|
4KUD
 
 | | Crystal structure of N-terminal acetylated Sir3 BAH domain D205N mutant in complex with yeast nucleosome core particle | | 分子名称: | Histone H2A.2, Histone H2B.1, Histone H3, ... | | 著者 | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | 登録日 | 2013-05-22 | | 公開日 | 2013-08-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.203 Å) | | 主引用文献 | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
7LPK
 
 | |
7LP0
 
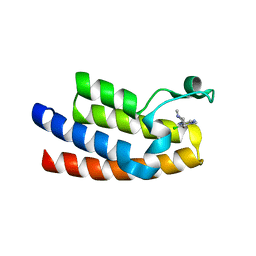 | |
7LRK
 
 | |
7LRO
 
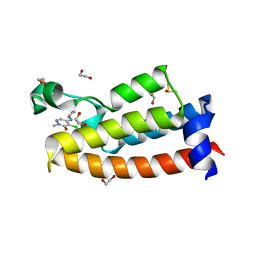 | |
2IW5
 
 | | Structural Basis for CoREST-Dependent Demethylation of Nucleosomes by the Human LSD1 Histone Demethylase | | 分子名称: | AMMONIUM ION, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Yang, M, Gocke, C.B, Luo, X, Borek, D, Tomchick, D.R, Machius, M, Otwinowski, Z, Yu, H. | | 登録日 | 2006-06-26 | | 公開日 | 2006-08-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Structural Basis for Corest-Dependent Demethylation of Nucleosomes by the Human Lsd1 Histone Demethylase
Mol.Cell, 23, 2006
|
|
7WLP
 
 | |
5FIU
 
 | | Binding and structural studies of a 5,5-difluoromethyl adenosine nucleoside with the fluorinase enzyme | | 分子名称: | 5'-FLUORO-5'-DEOXY-ADENOSINE SYNTHASE, 5,5-DIFLUOROMETHYL ADENOSINE, L(+)-TARTARIC ACID | | 著者 | Thompson, S, McMahon, S.A, Naismith, J.H, O'Hagan, D. | | 登録日 | 2015-10-02 | | 公開日 | 2015-12-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Exploration of a Potential Difluoromethyl-Nucleoside Substrate with the Fluorinase Enzyme.
Bioorg.Chem., 64, 2015
|
|
2Z83
 
 | |
7SO6
 
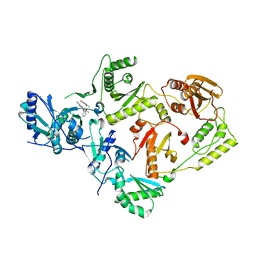 | | Crystal Structure of HIV-1 K103N, Y181C mutant Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-7-fluoro-2-naphthonitrile (JLJ635), a Non-nucleoside Inhibitor | | 分子名称: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-7-fluoronaphthalene-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | 著者 | Bertoletti, N, Frey, K.M, Anderson, K.S, Cisneros Trigo, J.A, Jorgensen, W.L, Chan, A.H. | | 登録日 | 2021-10-29 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7SO4
 
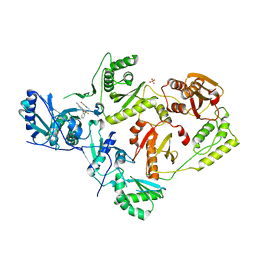 | | Crystal Structure of HIV-1 Y181C mutant Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-7-fluoro-2-naphthonitrile (JLJ635), a Non-nucleoside Inhibitor | | 分子名称: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-7-fluoronaphthalene-2-carbonitrile, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | 著者 | Bertoletti, N, Anderson, K.S, Cisneros Trigo, J.A, Jorgensen, W.L, Frey, K.M, Chan, A.H. | | 登録日 | 2021-10-29 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
5E5A
 
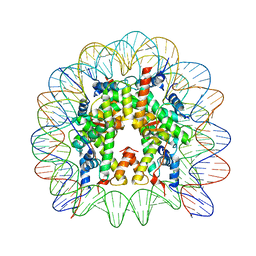 | | Crystal structure of the chromatin-tethering domain of Human cytomegalovirus IE1 protein bound to the nucleosome core particle | | 分子名称: | C-terminal domain of Regulatory protein IE1, DNA (146-MER), Histone H2A, ... | | 著者 | Fang, Q, Chen, P, Wang, M, Fang, J, Yang, N, Li, G, Xu, R.M. | | 登録日 | 2015-10-08 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.809 Å) | | 主引用文献 | Human cytomegalovirus IE1 protein alters the higher-order chromatin structure by targeting the acidic patch of the nucleosome
Elife, 5, 2016
|
|
6FTF
 
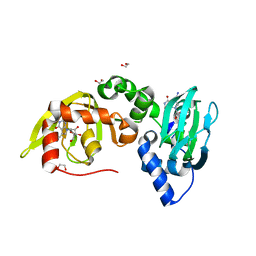 | | Regulatory subunit of a cAMP-independent protein kinase A from Trypanosoma cruzi at 1.09 A resolution | | 分子名称: | 1,2-ETHANEDIOL, 7-cyano-7-deazainosine, Protein kinase A regulatory subunit, ... | | 著者 | Volpato Santos, Y, Lorentzen, E, Basquin, J, Boshart, M. | | 登録日 | 2018-02-21 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.09151936 Å) | | 主引用文献 | Nucleoside analogue activators of cyclic AMP-independent protein kinase A of Trypanosoma.
Nat Commun, 10, 2019
|
|
2M11
 
 | | Structure of perimidinone-derived synthetic nucleoside paired with guanine in DNA duplex | | 分子名称: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(D3N)P*GP*CP*G)-3') | | 著者 | Kowal, E.A, Lad, R, Pallan, P.S, Muffly, E, Wawrzak, Z, Egli, M, Sturla, S.J, Stone, M.P. | | 登録日 | 2012-11-09 | | 公開日 | 2013-06-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Recognition of O6-benzyl-2'-deoxyguanosine by a perimidinone-derived synthetic nucleoside: a DNA interstrand stacking interaction.
Nucleic Acids Res., 41, 2013
|
|
8SPS
 
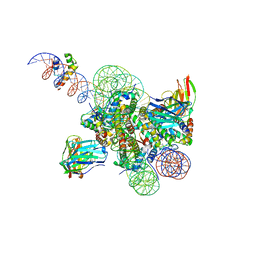 | | High resolution structure of ESRRB nucleosome bound OCT4 at site a and site b | | 分子名称: | DNA (168-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | 著者 | Lian, T, Guan, R, Bai, Y. | | 登録日 | 2023-05-03 | | 公開日 | 2023-06-28 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural mechanism of LIN28B nucleosome targeting by OCT4.
Mol.Cell, 83, 2023
|
|
8SPU
 
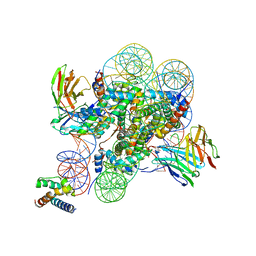 | | Structure of ESRRB nucleosome bound OCT4 at site c | | 分子名称: | DNA (168-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | 著者 | Lian, T, Guan, R, Bai, Y. | | 登録日 | 2023-05-03 | | 公開日 | 2023-06-28 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural mechanism of LIN28B nucleosome targeting by OCT4.
Mol.Cell, 83, 2023
|
|
4XHM
 
 | |
3A6N
 
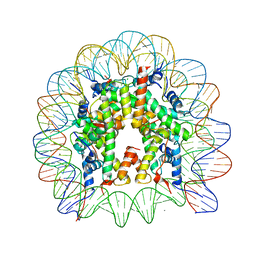 | | The nucleosome containing a testis-specific histone variant, human H3T | | 分子名称: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | 著者 | Tachiwana, H, Kagawa, W, Osakabe, A, Koichiro, K, Shiga, T, Kimura, H, Kurumizaka, H. | | 登録日 | 2009-09-04 | | 公開日 | 2010-05-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis of instability of the nucleosome containing a testis-specific histone variant, human H3T
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5L2B
 
 | |
5L24
 
 | |
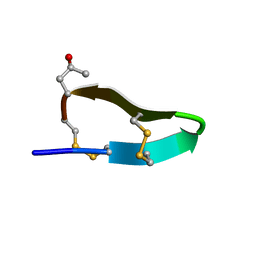
2M62
 
 | |
3AFA
 
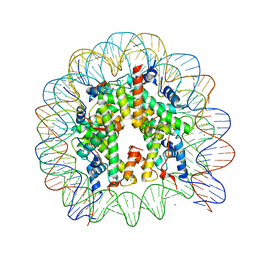 | | The human nucleosome structure | | 分子名称: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | 著者 | Tachiwana, H, Kagawa, W, Osakabe, A, Koichiro, K, Shiga, T, Kimura, H, Kurumizaka, H. | | 登録日 | 2010-02-24 | | 公開日 | 2010-05-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis of instability of the nucleosome containing a testis-specific histone variant, human H3T
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
