6HCF
 
 | | Structure of the rabbit 80S ribosome stalled on globin mRNA at the stop codon | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | 著者 | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | 登録日 | 2018-08-14 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
6HCJ
 
 | | Structure of the rabbit 80S ribosome on globin mRNA in the rotated state with A/P and P/E tRNAs | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | 著者 | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | 登録日 | 2018-08-15 | | 公開日 | 2018-10-17 | | 最終更新日 | 2018-11-14 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
6HCM
 
 | | Structure of the rabbit collided di-ribosome (stalled monosome) | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | 著者 | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | 登録日 | 2018-08-15 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (6.8 Å) | | 主引用文献 | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
6HCQ
 
 | | Structure of the rabbit collided di-ribosome (collided monosome) | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | 著者 | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | 登録日 | 2018-08-16 | | 公開日 | 2018-10-17 | | 最終更新日 | 2018-11-14 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
6HHQ
 
 | | Crystal structure of compound C45 bound to the yeast 80S ribosome | | 分子名称: | (3~{R})-3-[(1~{S})-2-[(1~{S},4~{a}~{R},6~{S},7~{S},8~{a}~{R})-6,7-bis(chloranyl)-5,5,8~{a}-trimethyl-2-methylidene-3,4,4~{a},6,7,8-hexahydro-1~{H}-naphthalen-1-yl]-1-oxidanyl-ethyl]pyrrolidine-2,5-dione, 18S ribosomal RNA, 25S ribosomal RNA, ... | | 著者 | Pellegrino, S, Vanderwal, C.D, Yusupov, M. | | 登録日 | 2018-08-28 | | 公開日 | 2019-02-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (3.10000038 Å) | | 主引用文献 | Understanding the role of intermolecular interactions between lissoclimides and the eukaryotic ribosome.
Nucleic Acids Res., 47, 2019
|
|
6HOG
 
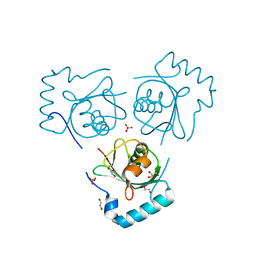 | | Structure of VPS34 LIR motif bound to GABARAP | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3,Gamma-aminobutyric acid receptor-associated protein, ... | | 著者 | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | 登録日 | 2018-09-17 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6HOH
 
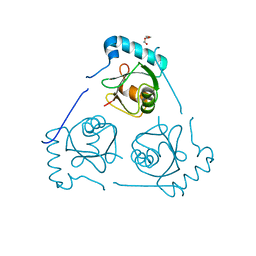 | | Structure of VPS34 LIR motif (S249E) bound to GABARAP | | 分子名称: | Phosphatidylinositol 3-kinase catalytic subunit type 3,Gamma-aminobutyric acid receptor-associated protein, TRIETHYLENE GLYCOL | | 著者 | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | 登録日 | 2018-09-17 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6HOI
 
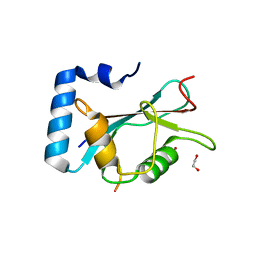 | | Structure of Beclin1 LIR motif bound to GABARAPL1 | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Beclin-1, ... | | 著者 | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | 登録日 | 2018-09-17 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.14 Å) | | 主引用文献 | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6HOJ
 
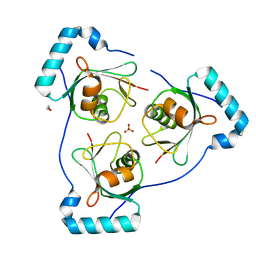 | | Structure of Beclin1 LIR motif bound to GABARAP | | 分子名称: | 1,2-ETHANEDIOL, Beclin-1,Gamma-aminobutyric acid receptor-associated protein, SULFATE ION | | 著者 | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | 登録日 | 2018-09-17 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6HOK
 
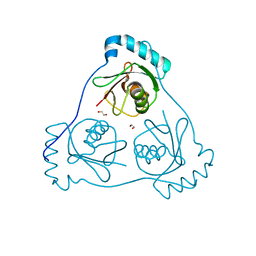 | | Structure of Beclin1 LIR (S96E) motif bound to GABARAP | | 分子名称: | 1,2-ETHANEDIOL, Beclin-1,Gamma-aminobutyric acid receptor-associated protein | | 著者 | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | 登録日 | 2018-09-17 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6HOL
 
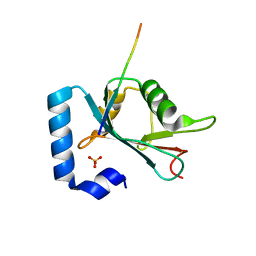 | | Structure of ATG14 LIR motif bound to GABARAPL1 | | 分子名称: | Beclin 1-associated autophagy-related key regulator, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein-like 1, ... | | 著者 | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | 登録日 | 2018-09-17 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6HR1
 
 | | Crystal structure of the YFPnano fusion protein | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | 著者 | Benoit, R.M. | | 登録日 | 2018-09-26 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Chimeric single alpha-helical domains as rigid fusion protein connections for protein nanotechnology and structural biology.
Structure, 2021
|
|
6HUT
 
 | |
6HWP
 
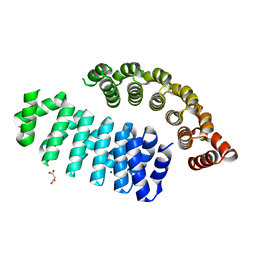 | |
6I7O
 
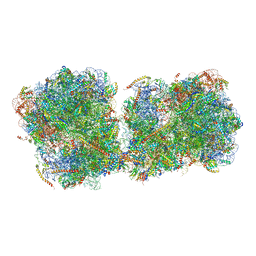 | | The structure of a di-ribosome (disome) as a unit for RQC and NGD quality control pathways recognition. | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Tesina, P, Cheng, J, Becker, T, Beckmann, R. | | 登録日 | 2018-11-16 | | 公開日 | 2019-01-16 | | 最終更新日 | 2019-03-13 | | 実験手法 | ELECTRON MICROSCOPY (5.3 Å) | | 主引用文献 | Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways.
EMBO J., 38, 2019
|
|
6IP5
 
 | | Cryo-EM structure of the CMV-stalled human 80S ribosome (Structure ii) | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Yokoyama, T, Shigematsu, H, Shirouzu, M, Imataka, H, Ito, T. | | 登録日 | 2018-11-02 | | 公開日 | 2019-05-29 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | HCV IRES Captures an Actively Translating 80S Ribosome.
Mol.Cell, 74, 2019
|
|
6IP6
 
 | | Cryo-EM structure of the CMV-stalled human 80S ribosome with HCV IRES (Structure iii) | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Yokoyama, T, Shigematsu, H, Shirouzu, M, Imataka, H, Ito, T. | | 登録日 | 2018-11-02 | | 公開日 | 2019-05-29 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | HCV IRES Captures an Actively Translating 80S Ribosome.
Mol.Cell, 74, 2019
|
|
6IP8
 
 | | Cryo-EM structure of the HCV IRES dependently initiated CMV-stalled 80S ribosome (Structure iv) | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Yokoyama, T, Shigematsu, H, Shirouzu, M, Imataka, H, Ito, T. | | 登録日 | 2018-11-02 | | 公開日 | 2019-05-29 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | HCV IRES Captures an Actively Translating 80S Ribosome.
Mol.Cell, 74, 2019
|
|
6IR1
 
 | | Crystal structure of red fluorescent protein mCherry complexed with the nanobody LaM4 at 1.9 Angstron resolution | | 分子名称: | MCherry fluorescent protein, mCherry's nanobody LaM4 | | 著者 | Ding, Y, Wang, Z.Y, Hu, R.T, Chen, X. | | 登録日 | 2018-11-09 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.919 Å) | | 主引用文献 | Structural insights into the binding of nanobodies LaM2 and LaM4 to the red fluorescent protein mCherry.
Protein Sci., 30, 2021
|
|
6IR2
 
 | | Crystal structure of red fluorescent protein mCherry complexed with the nanobody LaM2 at 1.4 Angstron resolution | | 分子名称: | MCherry fluorescent protein, mCherry's nanobody LaM2 | | 著者 | Ding, Y, Wang, Z.Y, Hu, R.T, Chen, X. | | 登録日 | 2018-11-09 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.393 Å) | | 主引用文献 | Structural insights into the binding of nanobodies LaM2 and LaM4 to the red fluorescent protein mCherry.
Protein Sci., 30, 2021
|
|
6IR6
 
 | | Green fluorescent protein variant GFPuv with the native lysine residue at the C-terminus | | 分子名称: | Green fluorescent protein, SULFATE ION | | 著者 | Nakatani, T, Yasui, N, Yamashita, A. | | 登録日 | 2018-11-12 | | 公開日 | 2019-04-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.642 Å) | | 主引用文献 | Specific modification at the C-terminal lysine residue of the green fluorescent protein variant, GFPuv, expressed in Escherichia coli.
Sci Rep, 9, 2019
|
|
6IR7
 
 | | Green fluorescent protein variant GFPuv with the modification to 6-hydroxynorleucine at the C-terminus | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-HYDROXY-L-NORLEUCINE, Green fluorescent protein, ... | | 著者 | Nakatani, T, Yasui, N, Yamashita, A. | | 登録日 | 2018-11-12 | | 公開日 | 2019-04-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.277 Å) | | 主引用文献 | Specific modification at the C-terminal lysine residue of the green fluorescent protein variant, GFPuv, expressed in Escherichia coli.
Sci Rep, 9, 2019
|
|
6ITC
 
 | | Structure of a substrate engaged SecA-SecY protein translocation machine | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | 著者 | Ma, C.Y, Wu, X.F, Sun, D.J, Park, E.Y, Rapoport, T.A, Gao, N, Long, L. | | 登録日 | 2018-11-21 | | 公開日 | 2019-06-12 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3.45 Å) | | 主引用文献 | Structure of the substrate-engaged SecA-SecY protein translocation machine.
Nat Commun, 10, 2019
|
|
6JGH
 
 | | Crystal structure of the F99S/M153T/V163A/T203I variant of GFP at 0.94 A | | 分子名称: | CHLORIDE ION, Green fluorescent protein | | 著者 | Eki, H, Tai, Y, Takaba, K, Hanazono, Y, Miki, K, Takeda, K. | | 登録日 | 2019-02-14 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (0.94 Å) | | 主引用文献 | Subatomic resolution X-ray structures of green fluorescent protein.
Iucrj, 6, 2019
|
|
6JGI
 
 | | Crystal structure of the S65T/F99S/M153T/V163A variant of GFP at 0.85 A | | 分子名称: | Green fluorescent protein | | 著者 | Tai, Y, Takaba, K, Hanazono, Y, Miki, K, Takeda, K. | | 登録日 | 2019-02-14 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Subatomic resolution X-ray structures of green fluorescent protein.
Iucrj, 6, 2019
|
|
