8FP4
 
 | |
8FPS
 
 | | GluA2 flip Q isoform N619K mutant of AMPA receptor in complex with gain-of-function TARP gamma-2, with 10mM CaCl2, 150mM NaCl, 1mM MgCl2, 330uM CTZ, and 100mM glutamate (Open-CaNaMg/N619K) | | 分子名称: | CHLORIDE ION, Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | 著者 | Nakagawa, T. | | 登録日 | 2023-01-05 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (2.38 Å) | | 主引用文献 | The open gate of the AMPA receptor forms a Ca 2+ binding site critical in regulating ion transport.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FP9
 
 | | GluA2 flip Q isoform of AMPA receptor in complex with gain-of-function TARP gamma-2, with 10mM CaCl2, 150mM NaCl, 1mM MgCl2, 330uM CTZ, and 100mM glutamate (Open-CaNaMg) | | 分子名称: | CALCIUM ION, CHLORIDE ION, Glutamate receptor 2, ... | | 著者 | Nakagawa, T. | | 登録日 | 2023-01-04 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.44 Å) | | 主引用文献 | The open gate of the AMPA receptor forms a Ca 2+ binding site critical in regulating ion transport.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FQF
 
 | |
8X93
 
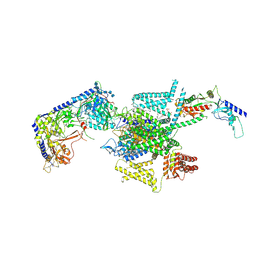 | | P/Q type calcium channel in complex with omega-Agatoxin IVA | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Yan, N, Li, Z, Cong, Y, Wu, T, Wang, T. | | 登録日 | 2023-11-29 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (2.92 Å) | | 主引用文献 | Structural basis for different omega-agatoxin IVA sensitivities of the P-type and Q-type Ca v 2.1 channels.
Cell Res., 34, 2024
|
|
8FPG
 
 | | GluA2 flip Q isoform of AMPA receptor in complex with gain-of-function TARP gamma-2, with 10mM CaCl2, 150mM NaCl, 1mM MgCl2, 330uM CTZ, and 100uM CNQX (Closed-CaNaMg) | | 分子名称: | CHLORIDE ION, Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | 著者 | Nakagawa, T. | | 登録日 | 2023-01-04 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (2.32 Å) | | 主引用文献 | The open gate of the AMPA receptor forms a Ca 2+ binding site critical in regulating ion transport.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FQ1
 
 | |
8FQB
 
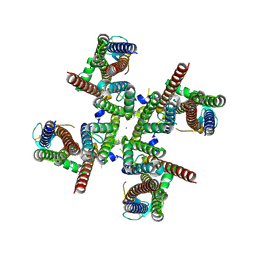 | | GluA2 flip Q isoform of AMPA receptor in complex with gain-of-function TARP gamma2, with 10mM CaCl2, 140mM NMDG, 330uM CTZ, and 100mM L-glutamate (Open-Ca10) | | 分子名称: | CALCIUM ION, CHLORIDE ION, Glutamate receptor 2, ... | | 著者 | Nakagawa, T. | | 登録日 | 2023-01-05 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (2.36 Å) | | 主引用文献 | The open gate of the AMPA receptor forms a Ca 2+ binding site critical in regulating ion transport.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FQ5
 
 | |
8PK0
 
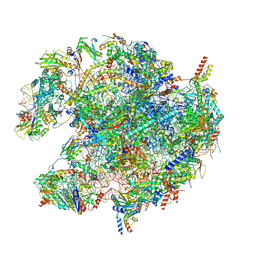 | | human mitoribosomal large subunit assembly intermediate 1 with GTPBP10-GTPBP7 | | 分子名称: | 16S rRNA + pre-H68-71 segment, 39S ribosomal protein L10, mitochondrial, ... | | 著者 | Kummer, E, Nguyen, T.G, Ritter, C. | | 登録日 | 2023-06-23 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | Structural insights into the role of GTPBP10 in the RNA maturation of the mitoribosome.
Nat Commun, 14, 2023
|
|
8PHJ
 
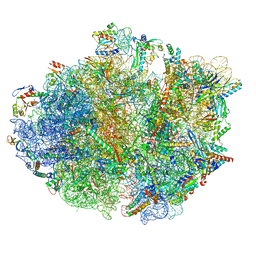 | | cA4-bound Cami1 in complex with 70S ribosome | | 分子名称: | 16S rRNA, 23S rRNA (2862-MER), 5S rRNA, ... | | 著者 | Tamulaitiene, G, Mogila, I, Sasnauskas, G, Tamulaitis, G. | | 登録日 | 2023-06-20 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.67 Å) | | 主引用文献 | Ribosomal stalk-captured CARF-RelE ribonuclease inhibits translation following CRISPR signaling.
Science, 382, 2023
|
|
5ZI3
 
 | | MDH3 wild type, apo-form | | 分子名称: | GLYCEROL, Malate dehydrogenase | | 著者 | Moriyama, S, Nishio, K, Mizushima, T. | | 登録日 | 2018-03-14 | | 公開日 | 2018-10-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of glyoxysomal malate dehydrogenase (MDH3) from Saccharomyces cerevisiae.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
8WKU
 
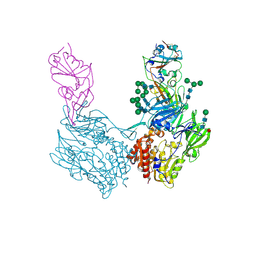 | | Complex structure of MjHKU4r-CoV-1 spike RBD bound to human CD26 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 soluble form, MjHKU4r-CoV-1 spike RBD, ... | | 著者 | Zhao, Z.N, Chai, Y, Gao, F. | | 登録日 | 2023-09-28 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Molecular basis for receptor recognition and broad host tropism for merbecovirus MjHKU4r-CoV-1.
Embo Rep., 25, 2024
|
|
8GIN
 
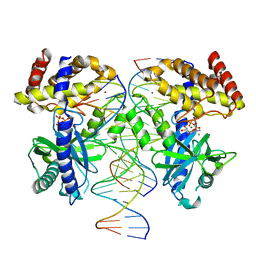 | |
8Q7C
 
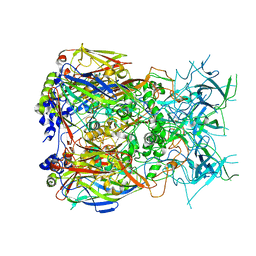 | |
5ZO2
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain in complex with mouse nectin-like molecule 1 (mNecl-1) Ig1 domain, 3.3A | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 3, Cell adhesion molecule 4 | | 著者 | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | 登録日 | 2018-04-12 | | 公開日 | 2019-01-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.29 Å) | | 主引用文献 | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8GUB
 
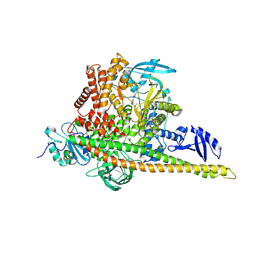 | | Cryo-EM structure of cancer-specific PI3Kalpha mutant H1047R in complex with BYL-719 | | 分子名称: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Liu, X, Zhou, Q, Hart, J.R, Xu, Y, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | 登録日 | 2022-09-11 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.73 Å) | | 主引用文献 | Cryo-EM structures of cancer-specific helical and kinase domain mutations of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8Q3Q
 
 | |
5ZI4
 
 | | MDH3 wild type, nad-oaa-form | | 分子名称: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXALOACETATE ION | | 著者 | Moriyama, S, Nishio, K, Mizushima, T. | | 登録日 | 2018-03-14 | | 公開日 | 2018-10-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of glyoxysomal malate dehydrogenase (MDH3) from Saccharomyces cerevisiae.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
8Q3N
 
 | |
8Q3P
 
 | |
8GNJ
 
 | |
8GQ5
 
 | |
8GNI
 
 | |
8GPN
 
 | | Human menin in complex with H3K79Me2 nucleosome | | 分子名称: | DNA (177-MER), Histone H2A type 1, Histone H2B 1.1, ... | | 著者 | Lin, J, Yu, D, Lam, W.H, Dang, S, Zhai, Y, Li, X.D. | | 登録日 | 2022-08-26 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-03-01 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Menin "reads" H3K79me2 mark in a nucleosomal context.
Science, 379, 2023
|
|
