5LEM
 
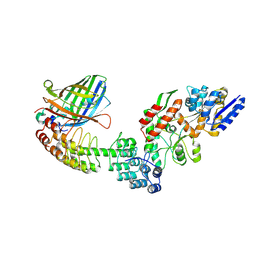 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_Off7_11_3G124 in complex with Maltose-binding Protein and Green Fluorescent Protein | | 分子名称: | DD_Off7_11_3G124, Green fluorescent protein, Maltose-binding periplasmic protein | | 著者 | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | 登録日 | 2016-06-30 | | 公開日 | 2017-08-02 | | 最終更新日 | 2019-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5LEL
 
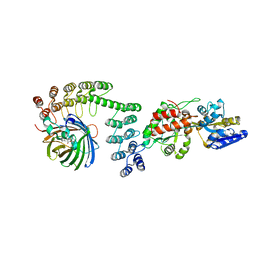 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_Off7_10_3G124 in complex with Maltose-binding Protein and Green Fluorescent Protein | | 分子名称: | DD_Off7_10_3G124, Green fluorescent protein, Maltose-binding periplasmic protein | | 著者 | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | 登録日 | 2016-06-30 | | 公開日 | 2017-11-15 | | 最終更新日 | 2019-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5GJO
 
 | |
5GJM
 
 | |
5GJP
 
 | |
5GJN
 
 | |
5KTG
 
 | |
5LKS
 
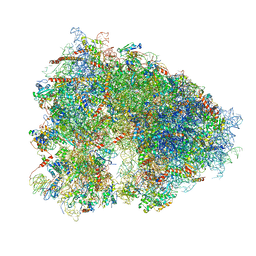 | | Structure-function insights reveal the human ribosome as a cancer target for antibiotics | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | 著者 | Myasnikov, A.G, Natchiar, S.K, Nebout, M, Hazemann, I, Imbert, V, Khatter, H, Peyron, J.-F, Klaholz, B.P. | | 登録日 | 2016-07-23 | | 公開日 | 2017-04-26 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure-function insights reveal the human ribosome as a cancer target for antibiotics.
Nat Commun, 7, 2016
|
|
5LL6
 
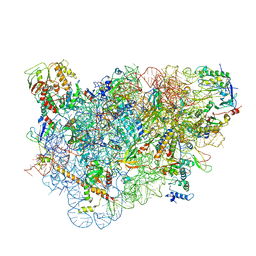 | | Structure of the 40S ABCE1 post-splitting complex in ribosome recycling and translation initiation | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | 著者 | Heuer, A, Gerovac, M, Schmidt, C, Trowitzsch, S, Preis, A, Koetter, P, Berninghausen, O, Becker, T, Beckmann, R, Tampe, R. | | 登録日 | 2016-07-26 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structure of the 40S-ABCE1 post-splitting complex in ribosome recycling and translation initiation.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5LOD
 
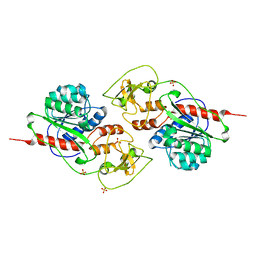 | |
5BJO
 
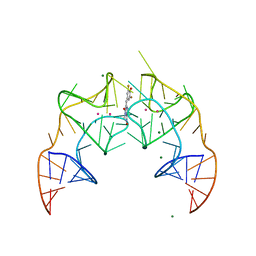 | | Crystal structure of the Corn RNA aptamer in complex with DFHO, site-specific 5-iodo-U | | 分子名称: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, MAGNESIUM ION, POTASSIUM ION, ... | | 著者 | Warner, K.D, Song, W, Filonov, G.S, Jaffrey, S.R, Ferre-D'Amare, A.R. | | 登録日 | 2016-08-21 | | 公開日 | 2017-09-27 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | A homodimer interface without base pairs in an RNA mimic of red fluorescent protein.
Nat. Chem. Biol., 13, 2017
|
|
5BJP
 
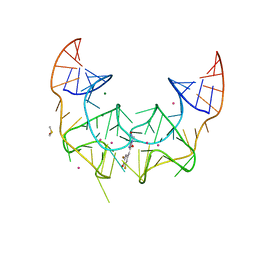 | | Crystal structure of the Corn RNA aptamer in complex with DFHO, iridium hexammine soak | | 分子名称: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, DIMETHYL SULFOXIDE, IRIDIUM ION, ... | | 著者 | Warner, K.D, Song, W, Filonov, G.S, Jaffrey, S.R, Ferre-D'Amare, A.R. | | 登録日 | 2016-08-21 | | 公開日 | 2017-09-27 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | A homodimer interface without base pairs in an RNA mimic of red fluorescent protein.
Nat. Chem. Biol., 13, 2017
|
|
5T2A
 
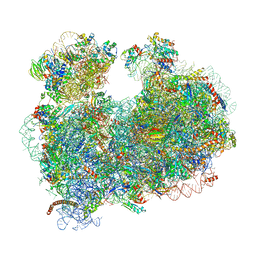 | | CryoEM structure of the Leishmania donovani 80S ribosome at 2.9 Angstrom resolution | | 分子名称: | 18S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Zhang, X, Lai, M, Zhou, Z.H. | | 登録日 | 2016-08-23 | | 公開日 | 2017-01-25 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structures and stabilization of kinetoplastid-specific split rRNAs revealed by comparing leishmanial and human ribosomes.
Nat Commun, 7, 2016
|
|
5T2C
 
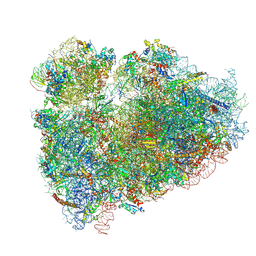 | | CryoEM structure of the human ribosome at 3.6 Angstrom resolution | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | 著者 | Zhang, X, Lai, M, Zhou, Z.H. | | 登録日 | 2016-08-23 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structures and stabilization of kinetoplastid-specific split rRNAs revealed by comparing leishmanial and human ribosomes.
Nat Commun, 7, 2016
|
|
5T3I
 
 | |
5T4I
 
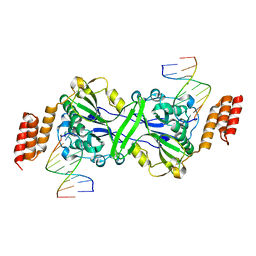 | |
5T5C
 
 | |
5LTP
 
 | |
5LTR
 
 | |
5LTQ
 
 | |
5LU3
 
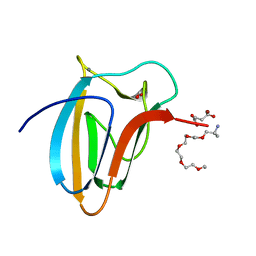 | | The Structure of Spirochaeta thermophila CBM64 | | 分子名称: | 3,6,9,12,15-pentaoxaoctadecan-17-amine, 4-oxobutanoic acid, CALCIUM ION, ... | | 著者 | Correia, M.A.S, Romao, M.J, Carvalho, A.L. | | 登録日 | 2016-09-07 | | 公開日 | 2017-02-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Stability and Ligand Promiscuity of Type A Carbohydrate-binding Modules Are Illustrated by the Structure of Spirochaeta thermophila StCBM64C.
J. Biol. Chem., 292, 2017
|
|
5GW4
 
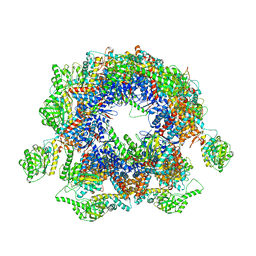 | | Structure of Yeast NPP-TRiC | | 分子名称: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | 著者 | Zang, Y, Jin, M, Wang, H, Cong, Y. | | 登録日 | 2016-09-08 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5GW5
 
 | | Structure of TRiC-AMP-PNP | | 分子名称: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | 著者 | Zang, Y, Jin, M, Wang, H, Cong, Y. | | 登録日 | 2016-09-08 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5TBW
 
 | | Crystal structure of chlorolissoclimide bound to the yeast 80S ribosome | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Konst, Z.A, Szklarski, A.R, Pellegrino, S, Michalak, S.E, Meyer, M, Zanette, C, Cencic, R, Nam, S, Horne, D.A, Pelletier, J, Mobley, D.L, Yusupova, G, Yusupov, M, Vanderwal, C.D. | | 登録日 | 2016-09-13 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Synthesis facilitates an understanding of the structural basis for translation inhibition by the lissoclimides.
Nat Chem, 9, 2017
|
|
5LYB
 
 | |
