3GJX
 
 | | Crystal Structure of the Nuclear Export Complex CRM1-Snurportin1-RanGTP | | 分子名称: | CHLORIDE ION, Exportin-1, GTP-binding nuclear protein Ran, ... | | 著者 | Monecke, T, Guettler, T, Neumann, P, Dickmanns, A, Goerlich, D, Ficner, R. | | 登録日 | 2009-03-09 | | 公開日 | 2009-05-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of the Nuclear Export Receptor CRM1 in Complex with Snurportin1 and RanGTP.
Science, 2009
|
|
8BPC
 
 | | Cryo-EM structure of the human SIN3B histone deacetylase core complex with SAHA at 2.8 Angstrom | | 分子名称: | CALCIUM ION, Histone deacetylase 2, Isoform 2 of Paired amphipathic helix protein Sin3b, ... | | 著者 | Wan, M.S.M, Muhammad, R, Koliopolous, M.G, Alfieri, C. | | 登録日 | 2022-11-16 | | 公開日 | 2023-05-10 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Mechanism of assembly, activation and lysine selection by the SIN3B histone deacetylase complex.
Nat Commun, 14, 2023
|
|
6OI3
 
 | | Crystal structure of human WDR5 in complex with monomethyl H3R2 peptide | | 分子名称: | GLYCEROL, Monomethyl H3R2 peptide, SULFATE ION, ... | | 著者 | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | 登録日 | 2019-04-08 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
6E49
 
 | | Pif1 peptide bound to PCNA trimer | | 分子名称: | ATP-dependent DNA helicase PIF1, Proliferating cell nuclear antigen | | 著者 | Buzovetsky, O, Kwon, Y, Pham, N.T, Kim, C, Ira, G, Sung, P, Xiong, Y. | | 登録日 | 2018-07-17 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Role of the Pif1-PCNA Complex in Pol delta-Dependent Strand Displacement DNA Synthesis and Break-Induced Replication.
Cell Rep, 21, 2017
|
|
6V8O
 
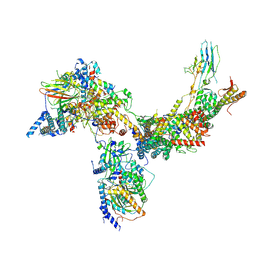 | | RSC core | | 分子名称: | Chromatin structure-remodeling complex protein RSC14, Chromatin structure-remodeling complex protein RSC3, Chromatin structure-remodeling complex protein RSC30, ... | | 著者 | Patel, A.B, Moore, C.M, Greber, B.J, Nogales, E. | | 登録日 | 2019-12-11 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement.
Elife, 8, 2019
|
|
6SFI
 
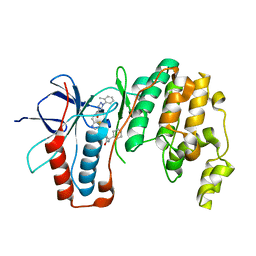 | | Crystal structure of p38 alpha in complex with compound 75 (MCP33) | | 分子名称: | Mitogen-activated protein kinase 14, ~{N}-[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]-2-[[[1-(2-methylphenyl)pyrazol-4-yl]carbonylamino]methyl]-1,3-thiazole-5-carboxamide | | 著者 | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2019-08-01 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6SFJ
 
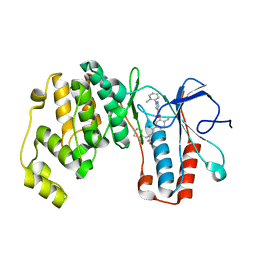 | | Crystal structure of p38 alpha in complex with compound 77 (MCP41) | | 分子名称: | Mitogen-activated protein kinase 14, ~{N}-[5-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]-2-methyl-phenyl]-1-(2-methylphenyl)pyrazole-4-carboxamide | | 著者 | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2019-08-01 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6SFK
 
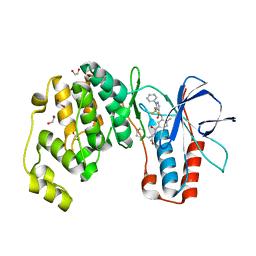 | | Crystal structure of p38 alpha in complex with compound 81 (MCP42) | | 分子名称: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 14, ~{N}-[5-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]-2-methyl-phenyl]-1-phenyl-5-(trifluoromethyl)pyrazole-4-carboxamide | | 著者 | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2019-08-01 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6SFO
 
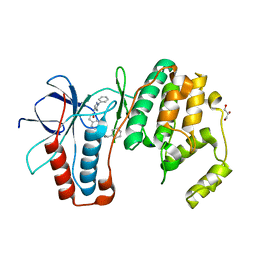 | | MAPK14 with bound inhibitor SR-318 | | 分子名称: | 5-azanyl-~{N}-[[4-(3-cyclohexylpropylcarbamoyl)phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, GLYCEROL, Mitogen-activated protein kinase 14, ... | | 著者 | Schroeder, M, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2019-08-01 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | MAPK14 with bound inhibitor SR-318
To Be Published
|
|
6PWW
 
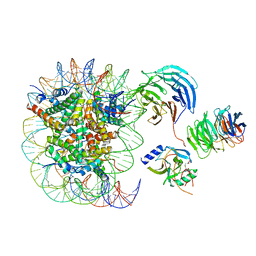 | | Cryo-EM structure of MLL1 in complex with RbBP5 and WDR5 bound to the nucleosome | | 分子名称: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | 著者 | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | 登録日 | 2019-07-23 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
8CVT
 
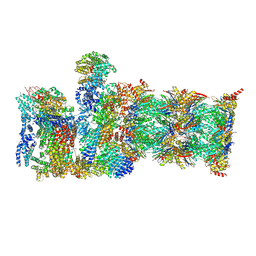 | | Human 19S-20S proteasome, state SD2 | | 分子名称: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 8, 26S proteasome complex subunit SEM1, ... | | 著者 | Zhao, J. | | 登録日 | 2022-05-18 | | 公開日 | 2022-11-02 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural insights into the human PA28-20S proteasome enabled by efficient tagging and purification of endogenous proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8JAL
 
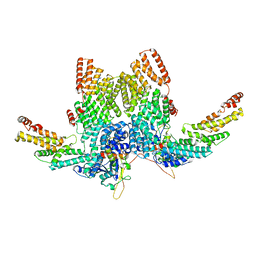 | | Structure of CRL2APPBP2 bound with RxxGP degron (dimer) | | 分子名称: | Amyloid protein-binding protein 2, Cullin-2, Elongin-B, ... | | 著者 | Zhao, S, Zhang, K, Xu, C. | | 登録日 | 2023-05-06 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAQ
 
 | | Structure of CRL2APPBP2 bound with RxxGP degron (tetramer) | | 分子名称: | Amyloid protein-binding protein 2, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Zhao, S, Zhang, K, Xu, C. | | 登録日 | 2023-05-06 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAS
 
 | | Structure of CRL2APPBP2 bound with RxxGPAA degron (tetramer) | | 分子名称: | Amyloid protein-binding protein 2, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Zhao, S, Zhang, K, Xu, C. | | 登録日 | 2023-05-07 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAV
 
 | | Structure of CRL2APPBP2 bound with the C-degron of MRPL28 (tetramer) | | 分子名称: | Amyloid protein-binding protein 2, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Zhao, S, Zhang, K, Xu, C. | | 登録日 | 2023-05-07 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | ELECTRON MICROSCOPY (3.44 Å) | | 主引用文献 | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3V81
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) with DNA and the nonnucleoside inhibitor nevirapine | | 分子名称: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3'), DNA (5'-D(*AP*TP*GP*GP*AP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), ... | | 著者 | Das, K, Martinez, S.E, Arnold, E. | | 登録日 | 2011-12-22 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.8503 Å) | | 主引用文献 | HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism.
Nat.Struct.Mol.Biol., 19, 2012
|
|
8JE2
 
 | | Cryo-EM structure of neddylated Cul2-Rbx1-EloBC-FEM1B complexed with FNIP1-FLCN | | 分子名称: | Cullin-2, Elongin-B, Elongin-C, ... | | 著者 | Dai, Z, Liang, L, Yin, Y.X. | | 登録日 | 2023-05-15 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.63 Å) | | 主引用文献 | Structural insights into the ubiquitylation strategy of the oligomeric CRL2 FEM1B E3 ubiquitin ligase.
Embo J., 43, 2024
|
|
6OR7
 
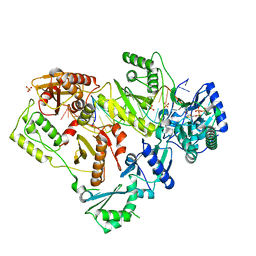 | | Structure of HIV-1 Reverse Transcriptase (RT) in complex with DNA AND (-)FTC-TP | | 分子名称: | DNA Primer 20-mer, DNA template 27-mer, MAGNESIUM ION, ... | | 著者 | Bertoletti, N, Chan, A.H, Anderson, K.S. | | 登録日 | 2019-04-29 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Structural insights into the recognition of nucleoside reverse transcriptase inhibitors by HIV-1 reverse transcriptase: First crystal structures with reverse transcriptase and the active triphosphate forms of lamivudine and emtricitabine.
Protein Sci., 28, 2019
|
|
2PP4
 
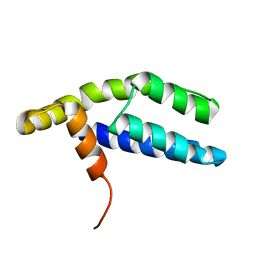 | | Solution Structure of ETO-TAFH refined in explicit solvent | | 分子名称: | Protein ETO | | 著者 | Wei, Y, Liu, S, Lausen, J, Woodrell, C, Cho, S, Biris, N, Kobayashi, N, Yokoyama, S, Werner, M.H. | | 登録日 | 2007-04-27 | | 公開日 | 2007-06-19 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A TAF4-homology domain from the corepressor ETO is a docking platform for positive and negative regulators of transcription
Nat.Struct.Mol.Biol., 14, 2007
|
|
6OUN
 
 | |
6OTZ
 
 | |
6P2G
 
 | | Structure of HIV-1 Reverse Transcriptase (RT) in complex with dsDNA and D-ddCTP | | 分子名称: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA Primer 20-mer, DNA template 27-mer, ... | | 著者 | Bertoletti, N, Anderson, K.S. | | 登録日 | 2019-05-21 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Structural insights into the recognition of nucleoside reverse transcriptase inhibitors by HIV-1 reverse transcriptase: First crystal structures with reverse transcriptase and the active triphosphate forms of lamivudine and emtricitabine.
Protein Sci., 28, 2019
|
|
6P1I
 
 | | Structure of HIV-1 Reverse Transcriptase (RT) in complex with dsDNA and dCTP | | 分子名称: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA Primer 20-mer, DNA template 27-mer, ... | | 著者 | Bertoletti, N, Anderson, K.S. | | 登録日 | 2019-05-19 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.74 Å) | | 主引用文献 | Structural insights into the recognition of nucleoside reverse transcriptase inhibitors by HIV-1 reverse transcriptase: First crystal structures with reverse transcriptase and the active triphosphate forms of lamivudine and emtricitabine.
Protein Sci., 28, 2019
|
|
6P1X
 
 | |
4LFI
 
 | | Crystal structure of scCK2 alpha in complex with GMPPNP | | 分子名称: | Casein kinase II subunit alpha, GLYCEROL, MANGANESE (II) ION, ... | | 著者 | Liu, H. | | 登録日 | 2013-06-27 | | 公開日 | 2014-03-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The multiple nucleotide-divalent cation binding modes of Saccharomyces cerevisiae CK2 alpha indicate a possible co-substrate hydrolysis product (ADP/GDP) release pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
