7XRY
 
 | | Crystal structure of MERS main protease in complex with inhibitor YH-53 | | 分子名称: | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, ORF1a | | 著者 | Lin, C, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | 登録日 | 2022-05-12 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structural Basis for the Inhibition of Coronaviral Main Proteases by a Benzothiazole-Based Inhibitor.
Viruses, 14, 2022
|
|
7XRS
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with inhibitor YH-53 | | 分子名称: | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, Replicase polyprotein 1a | | 著者 | Zhou, X.L, Zhong, F.L, Lin, C, Zeng, P, Zhang, J, Li, J. | | 登録日 | 2022-05-11 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structural Basis for the Inhibition of Coronaviral Main Proteases by a Benzothiazole-Based Inhibitor.
Viruses, 14, 2022
|
|
7YGQ
 
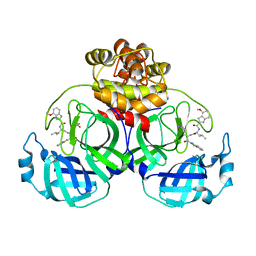 | | Crystal structure of SARS main protease in complex with inhibitor YH-53 | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Lin, C, Zhong, F.L, Zhou, X.L, Zeng, P, Zhang, J, Li, J. | | 登録日 | 2022-07-12 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structural Basis for the Inhibition of Coronaviral Main Proteases by a Benzothiazole-Based Inhibitor.
Viruses, 14, 2022
|
|
6AAA
 
 | |
4WBX
 
 | | Conserved hypothetical protein PF1771 from Pyrococcus furiosus solved by sulfur SAD using Swiss Light Source data | | 分子名称: | 2-keto acid:ferredoxin oxidoreductase subunit alpha | | 著者 | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2014-09-04 | | 公開日 | 2014-12-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
2RLQ
 
 | | NMR structure of CCP modules 2-3 of complement factor H | | 分子名称: | Complement factor H | | 著者 | Hocking, H.G, Herbert, A.P, Pangburn, M.K, Kavanagh, D, Barlow, P.N, Uhrin, D. | | 登録日 | 2007-07-29 | | 公開日 | 2008-02-19 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the N-terminal region of complement factor H and conformational implications of disease-linked sequence variations.
J.Biol.Chem., 283, 2008
|
|
3W02
 
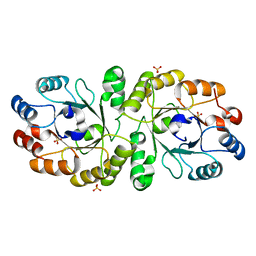 | | Crystal structure of PcrB complexed with SO4 from Staphylococcus aureus subsp. aureus Mu3 | | 分子名称: | Heptaprenylglyceryl phosphate synthase, SULFATE ION | | 著者 | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | 登録日 | 2012-10-17 | | 公開日 | 2012-12-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
6AHS
 
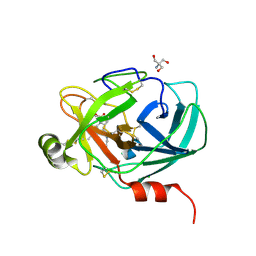 | | Mouse Kallikrein 7 in complex with imidazolinylindole derivative | | 分子名称: | 1-[(2-chlorophenyl)sulfonyl]-5-methyl-3-[(4R)-2-methyl-4,5-dihydro-1H-imidazol-4-yl]-1H-indole, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | 著者 | Sugawara, H. | | 登録日 | 2018-08-20 | | 公開日 | 2019-01-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Discovery and structure-activity relationship of imidazolinylindole derivatives as kallikrein 7 inhibitors.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
3W01
 
 | | Crystal structure of PcrB complexed with PEG from Staphylococcus aureus subsp. aureus Mu3 | | 分子名称: | Heptaprenylglyceryl phosphate synthase, TRIETHYLENE GLYCOL | | 著者 | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | 登録日 | 2012-10-17 | | 公開日 | 2012-12-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3VJI
 
 | | Human PPAR gamma ligand binding domain in complex with JKPL53 | | 分子名称: | (2S)-2-{4-butoxy-3-[({4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]benzoyl}amino)methyl]benzyl}butanoic acid, Peroxisome proliferator-activated receptor gamma | | 著者 | Tomioka, D, Kuwabara, N, Hashimoto, H, Sato, M, Shimizu, T. | | 登録日 | 2011-10-20 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Peroxisome proliferator-activated receptors (PPARs) have multiple binding points that accommodate ligands in various conformations: phenylpropanoic acid-type PPAR ligands bind to PPAR in different conformations, depending on the subtype.
J.Med.Chem., 55, 2012
|
|
3VSU
 
 | | The complex structure of XylC with xylobiose | | 分子名称: | Xylosidase, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | 著者 | Huang, C.H, Sun, Y, Ko, T.P, Ma, Y, Chen, C.C, Zheng, Y, Chan, H.C, Pang, X, Wiegel, J, Shao, W, Guo, R.T. | | 登録日 | 2012-05-09 | | 公開日 | 2013-02-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | The substrate/product-binding modes of a novel GH120 beta-xylosidase (XylC) from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Biochem.J., 448, 2012
|
|
3VI8
 
 | | Human PPAR alpha ligand binding domain in complex with a synthetic agonist APHM13 | | 分子名称: | (2S)-2-(4-methoxy-3-{[(pyren-1-ylcarbonyl)amino]methyl}benzyl)butanoic acid, Peroxisome proliferator-activated receptor alpha | | 著者 | Oyama, T, Miyachi, H, Morikawa, K. | | 登録日 | 2011-09-25 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Peroxisome proliferator-activated receptors (PPARs) have multiple binding points that accommodate ligands in various conformations: phenylpropanoic acid-type PPAR ligands bind to PPAR in different conformations, depending on the subtype
J.Med.Chem., 55, 2012
|
|
6O83
 
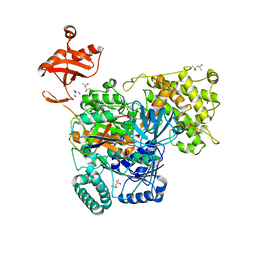 | | S. pombe ubiquitin E1~ubiquitin-AMP tetrahedral intermediate mimic | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-{[(3-aminopropyl)sulfonyl]amino}-5'-deoxyadenosine, ... | | 著者 | Hann, Z.S, Lima, C.D. | | 登録日 | 2019-03-08 | | 公開日 | 2019-06-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.153 Å) | | 主引用文献 | Structural basis for adenylation and thioester bond formation in the ubiquitin E1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5LE8
 
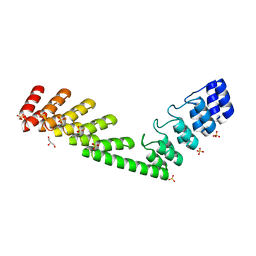 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_D12_15_D12 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DD_D12_15_D12, GLYCEROL, ... | | 著者 | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | 登録日 | 2016-06-29 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
3VZZ
 
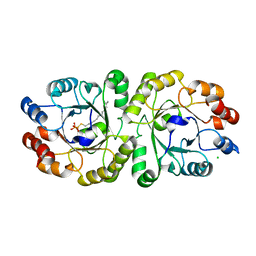 | | Crystal structure of PcrB complexed with FsPP from bacillus subtilis subap. subtilis str. 168 | | 分子名称: | CHLORIDE ION, Heptaprenylglyceryl phosphate synthase, MAGNESIUM ION, ... | | 著者 | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | 登録日 | 2012-10-17 | | 公開日 | 2012-12-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
1RNQ
 
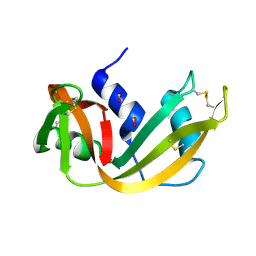 | | RIBONUCLEASE A CRYSTALLIZED FROM 8M SODIUM FORMATE | | 分子名称: | FORMIC ACID, RIBONUCLEASE A | | 著者 | Fedorov, A.A, Josef-Mccarthy, D, Graf, I, Anguelova, D, Fedorov, E.V, Almo, S.C. | | 登録日 | 1995-11-08 | | 公開日 | 1996-04-03 | | 最終更新日 | 2019-08-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Ionic interactions in crystalline bovine pancreatic ribonuclease A.
Biochemistry, 35, 1996
|
|
3WUM
 
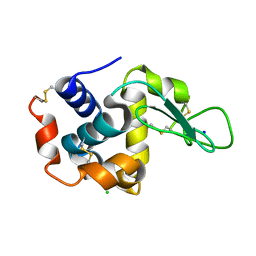 | |
2RLP
 
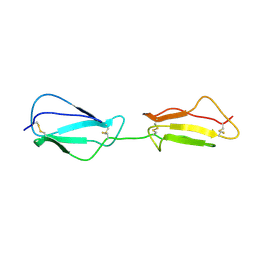 | | NMR structure of CCP modules 1-2 of complement factor H | | 分子名称: | Complement factor H | | 著者 | Hocking, H.G, Herbert, A.P, Pangburn, M.K, Kavanagh, D, Barlow, P.N, Uhrin, D. | | 登録日 | 2007-07-28 | | 公開日 | 2008-02-19 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the N-terminal region of complement factor H and conformational implications of disease-linked sequence variations.
J.Biol.Chem., 283, 2008
|
|
6EXW
 
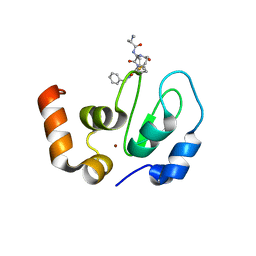 | | Crystal structure of cIAP1-BIR3 in complex with a covalently bound SM | | 分子名称: | (3~{S},6~{S},7~{R},9~{a}~{S})-6-[[(2~{S})-2-(methylamino)propanoyl]amino]-5-oxidanylidene-~{N}-(phenylmethyl)-7-[(propanoylamino)methyl]-3,6,7,8,9,9~{a}-hexahydropyrrolo[1,2-a]azepine-3-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | 著者 | Corti, A, Cossu, F, Milani, M, Mastrangelo, E. | | 登録日 | 2017-11-10 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure-based design and molecular profiling of Smac-mimetics selective for cellular IAPs.
FEBS J., 285, 2018
|
|
6ET9
 
 | | Structure of the acetoacetyl-CoA-thiolase/HMG-CoA-synthase complex from Methanothermococcus thermolithotrophicus at 2.75 A | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acetyl-CoA acetyltransferase thiolase, CHLORIDE ION, ... | | 著者 | Engilberge, S, Voegeli, B, Girard, E, Riobe, F, Maury, O, Erb, T.J, Shima, S, Wagner, T. | | 登録日 | 2017-10-25 | | 公開日 | 2018-03-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Archaeal acetoacetyl-CoA thiolase/HMG-CoA synthase complex channels the intermediate via a fused CoA-binding site.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3WXQ
 
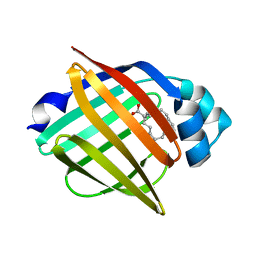 | | Serial femtosecond X-ray structure of human fatty acid-binding protein type-3 (FABP3) in complex with stearic acid (C18:0) determined using X-ray free-electron laser at SACLA | | 分子名称: | Fatty acid-binding protein, heart, STEARIC ACID | | 著者 | Mizohata, E, Suzuki, M, Kakinouchi, K, Sugiyama, S, Murata, M, Sugahara, M, Nango, E, Tanaka, T, Tanaka, R, Tono, K, Song, C, Hatsui, T, Joti, Y, Yabashi, M, Iwata, S. | | 登録日 | 2014-08-04 | | 公開日 | 2014-11-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat. Methods, 12, 2015
|
|
3WUL
 
 | |
6F4Q
 
 | | Human JMJD5 (Q275C) in complex with Mn(II), NOG and RPS6-A138C (129-144) (complex-2) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 40S ribosomal protein S6, GLYCEROL, ... | | 著者 | Chowdhury, R, Islam, M.S, Schofield, C.J. | | 登録日 | 2017-11-29 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | JMJD5 is a human arginyl C-3 hydroxylase.
Nat Commun, 9, 2018
|
|
3WXS
 
 | | Thaumatin structure determined by SPring-8 Angstrom Compact free electron Laser (SACLA) | | 分子名称: | L(+)-TARTARIC ACID, thaumatin I | | 著者 | Masuda, T, Nango, E, Sugahara, M, Mizohata, E, Tanaka, T, Tanaka, R, Suzuki, M, Mikami, B, Iwata, S. | | 登録日 | 2014-08-07 | | 公開日 | 2014-11-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat. Methods, 12, 2015
|
|
3WXT
 
 | |
