3W9G
 
 | |
4QQI
 
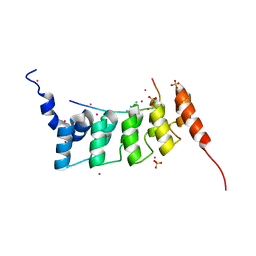 | | Crystal structure of ANKRA2-RFX7 complex | | 分子名称: | Ankyrin repeat family A protein 2, DNA-binding protein RFX7, SULFATE ION, ... | | 著者 | Xu, C, Tempel, W, Dong, A, Mackenzie, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2014-06-27 | | 公開日 | 2014-09-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Crystal structure of ANKRA2-RFX7 complex
To Be Published
|
|
4RLV
 
 | |
4RLY
 
 | |
4OT9
 
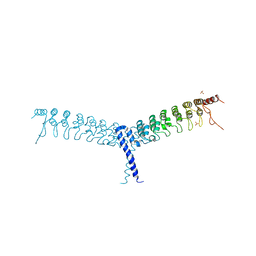 | | crystal structure of the C-terminal domain of p100/NF-kB2 | | 分子名称: | Nuclear factor NF-kappa-B p100 subunit, SULFATE ION | | 著者 | Tao, Z.H, Huang, D.B, Fusco, A, Gupta, K, Ware, C.F, Duynne, G.V. | | 登録日 | 2014-02-13 | | 公開日 | 2015-01-14 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | p100/I kappa B delta sequesters and inhibits NF-kappa B through kappaBsome formation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3J9P
 
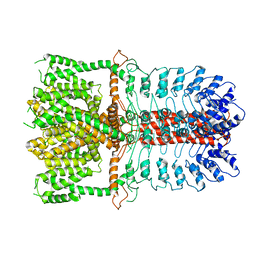 | | Structure of the TRPA1 ion channel determined by electron cryo-microscopy | | 分子名称: | Maltose-binding periplasmic protein, Transient receptor potential cation channel subfamily A member 1 chimera | | 著者 | Paulsen, C.E, Armache, J.-P, Gao, Y, Cheng, Y, Julius, D. | | 登録日 | 2015-02-14 | | 公開日 | 2015-04-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.24 Å) | | 主引用文献 | Structure of the TRPA1 ion channel suggests regulatory mechanisms.
Nature, 520, 2015
|
|
3J9J
 
 | | Structure of the capsaicin receptor, TRPV1, determined by single particle electron cryo-microscopy | | 分子名称: | Transient receptor potential cation channel subfamily V member 1 | | 著者 | Wang, R.Y.-R, Barad, B.A, Fraser, J.S, DiMaio, F. | | 登録日 | 2015-02-02 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.275 Å) | | 主引用文献 | EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy.
Nat.Methods, 12, 2015
|
|
5CZY
 
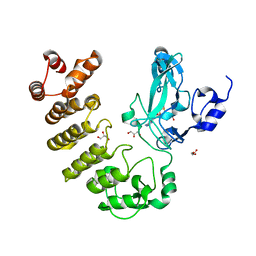 | | Crystal structure of LegAS4 | | 分子名称: | GLYCEROL, Legionella effector LegAS4, S-ADENOSYLMETHIONINE | | 著者 | Son, J, Hwang, K.Y, Lee, W.C. | | 登録日 | 2015-08-01 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of Legionella pneumophila type IV secretion system effector LegAS4
Biochem.Biophys.Res.Commun., 465, 2015
|
|
5D68
 
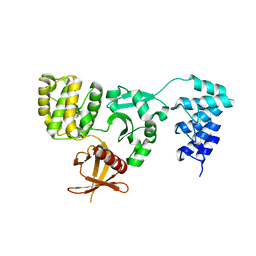 | | Crystal structure of KRIT1 ARD-FERM | | 分子名称: | Krev interaction trapped protein 1 | | 著者 | Zhang, R, Li, X, Boggon, T.J. | | 登録日 | 2015-08-11 | | 公開日 | 2015-10-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.908 Å) | | 主引用文献 | Structural analysis of the KRIT1 ankyrin repeat and FERM domains reveals a conformationally stable ARD-FERM interface.
J.Struct.Biol., 192, 2015
|
|
5IS0
 
 | | Structure of TRPV1 in complex with capsazepine, determined in lipid nanodisc | | 分子名称: | Transient receptor potential cation channel subfamily V member 1, capsazepine | | 著者 | Gao, Y, Cao, E, Julius, D, Cheng, Y. | | 登録日 | 2016-03-15 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.43 Å) | | 主引用文献 | TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action.
Nature, 534, 2016
|
|
5IRX
 
 | | Structure of TRPV1 in complex with DkTx and RTX, determined in lipid nanodisc | | 分子名称: | (2S)-2-(acetyloxy)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}propyl pentanoate, (2S)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexanoyloxy)propyl hexanoate, (4R,7S)-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-7-[(pentanoyloxy)methyl]-3,5,8-trioxa-4lambda~5~-phosphatetradecan-1-aminium, ... | | 著者 | Gao, Y, Cao, E, Julius, D, Cheng, Y. | | 登録日 | 2016-03-14 | | 公開日 | 2016-05-25 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action.
Nature, 534, 2016
|
|
5IRZ
 
 | | Structure of TRPV1 determined in lipid nanodisc | | 分子名称: | (2S)-1-{[(R)-hydroxy{[(1R,2R,3S,4S,5S,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-3-(pentanoyloxy)propan-2-yl decanoate, (2S)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexanoyloxy)propyl hexanoate, (4R,7S)-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-7-[(pentanoyloxy)methyl]-3,5,8-trioxa-4lambda~5~-phosphatetradecan-1-aminium, ... | | 著者 | Gao, Y, Cao, E, Julius, D, Cheng, Y. | | 登録日 | 2016-03-15 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action.
Nature, 534, 2016
|
|
5JA4
 
 | |
5AAR
 
 | |
5JHQ
 
 | |
5H2C
 
 | | Crystal structure of Saccharomyces cerevisiae Osh1 ANK - Nvj1 | | 分子名称: | Nucleus-vacuole junction protein 1, Oxysterol-binding protein homolog 1 | | 著者 | Im, Y.J, Manik, M.K, Yang, H.S, Tong, J.S. | | 登録日 | 2016-10-14 | | 公開日 | 2017-05-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.508 Å) | | 主引用文献 | Structure of Yeast OSBP-Related Protein Osh1 Reveals Key Determinants for Lipid Transport and Protein Targeting at the Nucleus-Vacuole Junction
Structure, 25, 2017
|
|
5H2A
 
 | |
5H28
 
 | |
5VKQ
 
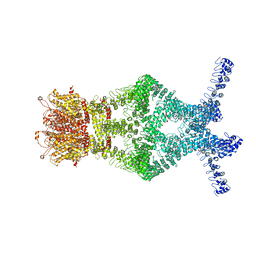 | | Structure of a mechanotransduction ion channel Drosophila NOMPC in nanodisc | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, No mechanoreceptor potential C isoform L | | 著者 | Jin, P, Bulkley, D, Guo, Y, Zhang, W, Guo, Z, Huynh, W, Wu, S, Meltzer, S, Chen, T, Jan, L.Y, Jan, Y.-N, Cheng, Y. | | 登録日 | 2017-04-22 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.55 Å) | | 主引用文献 | Electron cryo-microscopy structure of the mechanotransduction channel NOMPC.
Nature, 547, 2017
|
|
5GP7
 
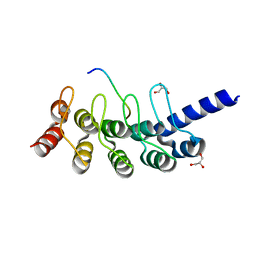 | | Structural basis for the binding between Tankyrase-1 and USP25 | | 分子名称: | GLYCEROL, Tankyrase-1, Ubiquitin carboxyl-terminal hydrolase 25 | | 著者 | Liu, J, Xu, D, Fu, T, Pan, L. | | 登録日 | 2016-08-01 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.502 Å) | | 主引用文献 | USP25 regulates Wnt signaling by controlling the stability of tankyrases
Genes Dev., 31, 2017
|
|
5Y4F
 
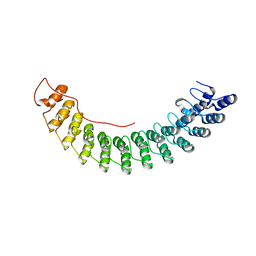 | | Crystal Structure of AnkB Ankyrin Repeats R13-24 in complex with autoinhibition segment AI-c | | 分子名称: | ACETATE ION, Ankyrin-2, CALCIUM ION | | 著者 | Chen, K, Li, J, Wang, C, Wei, Z, Zhang, M. | | 登録日 | 2017-08-03 | | 公開日 | 2017-09-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.953 Å) | | 主引用文献 | Autoinhibition of ankyrin-B/G membrane target bindings by intrinsically disordered segments from the tail regions.
Elife, 6, 2017
|
|
5Y4E
 
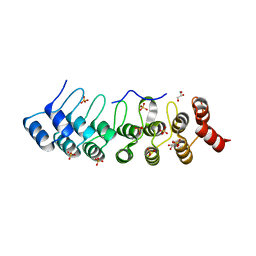 | | Crystal Structure of AnkB Ankyrin Repeats R8-14 in complex with autoinhibition segment AI-b | | 分子名称: | Ankyrin-2,Ankyrin-2, GLYCEROL, SULFATE ION | | 著者 | Chen, K, Li, J, Wang, C, Wei, Z, Zhang, M. | | 登録日 | 2017-08-03 | | 公開日 | 2017-09-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.341 Å) | | 主引用文献 | Autoinhibition of ankyrin-B/G membrane target bindings by intrinsically disordered segments from the tail regions.
Elife, 6, 2017
|
|
5Y4D
 
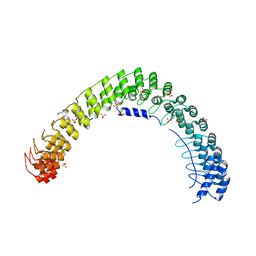 | | Crystal Structure of AnkB Ankyrin Repeats in Complex with AnkR/AnkB Chimeric Autoinhibition Segment | | 分子名称: | Ankyrin-1,Ankyrin-2,Ankyrin-2, SULFATE ION | | 著者 | Chen, K, Li, J, Wang, C, Wei, Z, Zhang, M. | | 登録日 | 2017-08-03 | | 公開日 | 2017-09-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Autoinhibition of ankyrin-B/G membrane target bindings by intrinsically disordered segments from the tail regions.
Elife, 6, 2017
|
|
5YBJ
 
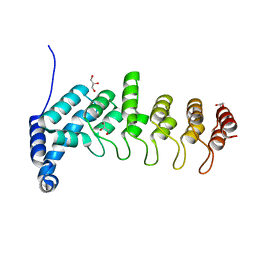 | | Structure of apo KANK1 ankyrin domain | | 分子名称: | GLYCEROL, KN motif and ankyrin repeat domain-containing protein 1 | | 著者 | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | 登録日 | 2017-09-05 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.341 Å) | | 主引用文献 | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
5YBU
 
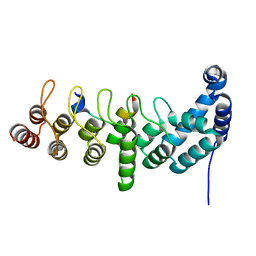 | | Structure of the KANK1 ankyrin domain in complex with KIF21A peptide | | 分子名称: | KN motif and ankyrin repeat domain-containing protein 1, Kinesin-like protein KIF21A | | 著者 | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | 登録日 | 2017-09-05 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
