5KDM
 
 | |
1SUQ
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R185545 | | 分子名称: | (6-[4-(AMINOMETHYL)-2,6-DIMETHYLPHENOXY]-2-{[4-(AMINOMETHYL)PHENYL]AMINO}-5-BROMOPYRIMIDIN-4-YL)METHANOL, MAGNESIUM ION, REVERSE TRANSCRIPTASE | | 著者 | Das, K, Arnold, E. | | 登録日 | 2004-03-26 | | 公開日 | 2004-05-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants
J.Med.Chem., 47, 2004
|
|
1SV5
 
 | | CRYSTAL STRUCTURE OF K103N MUTANT HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R165335 | | 分子名称: | 4-({6-AMINO-5-BROMO-2-[(4-CYANOPHENYL)AMINO]PYRIMIDIN-4-YL}OXY)-3,5-DIMETHYLBENZONITRILE, Reverse Transcriptase | | 著者 | Das, K, Arnold, E. | | 登録日 | 2004-03-27 | | 公開日 | 2004-05-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants
J.Med.Chem., 47, 2004
|
|
6I7B
 
 | | Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 3. | | 分子名称: | Nucleoprotein | | 著者 | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Carlero, D, Ortin, J, Martin-Benito, J. | | 登録日 | 2018-11-16 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (10 Å) | | 主引用文献 | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
5D3G
 
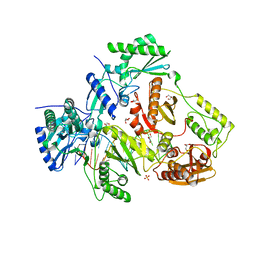 | | Structure of HIV-1 Reverse Transcriptase Bound to a Novel 38-mer Hairpin Template-Primer DNA Aptamer | | 分子名称: | DNA aptamer (38-MER), GLYCEROL, HIV-1 REVERSE TRANSCRIPTASE P51 subunit, ... | | 著者 | Miller, M.T, Tuske, S, Das, K, Arnold, E. | | 登録日 | 2015-08-06 | | 公開日 | 2015-09-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of HIV-1 reverse transcriptase bound to a novel 38-mer hairpin template-primer DNA aptamer.
Protein Sci., 25, 2016
|
|
8HXZ
 
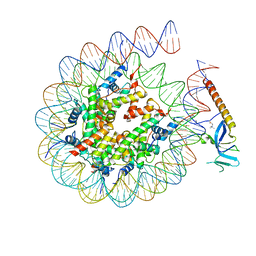 | |
8X15
 
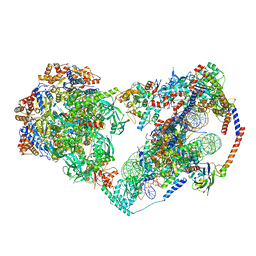 | | Structure of nucleosome-bound SRCAP-C in the apo state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | 著者 | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | 登録日 | 2023-11-06 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insights into histone exchange by human SRCAP complex.
Cell Discov, 10, 2024
|
|
8X1C
 
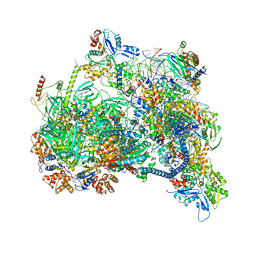 | | Structure of nucleosome-bound SRCAP-C in the ADP-bound state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | 著者 | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | 登録日 | 2023-11-06 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insights into histone exchange by human SRCAP complex.
Cell Discov, 10, 2024
|
|
8X19
 
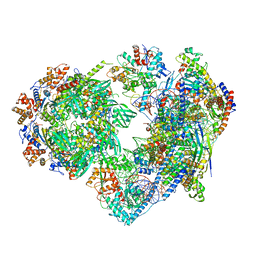 | | Structure of nucleosome-bound SRCAP-C in the ADP-BeFx-bound state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | 著者 | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | 登録日 | 2023-11-06 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insights into histone exchange by human SRCAP complex.
Cell Discov, 10, 2024
|
|
2I5W
 
 | |
7ZI4
 
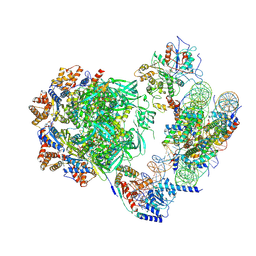 | | Cryo-EM structure of the human INO80 complex bound to a WT nucleosome | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, BERYLLIUM TRIFLUORIDE ION, ... | | 著者 | Vance, N.R, Ayala, R, Willhoft, O, Tvardovskiy, A, McCormack, E.A, Bartke, T, Zhang, X, Wigley, D.B. | | 登録日 | 2022-04-07 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structure of the human INO80 complex bound to a WT nucleosome
To Be Published
|
|
6IRO
 
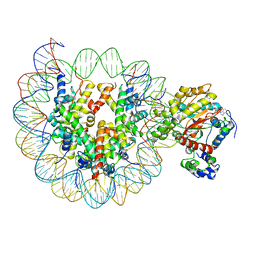 | | the crosslinked complex of ISWI-nucleosome in the ADP-bound state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (167-MER), Histone H2A, ... | | 著者 | Yan, L.J, Wu, H, Li, X.M, Gao, N, Chen, Z.C. | | 登録日 | 2018-11-13 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling.
Nat. Struct. Mol. Biol., 26, 2019
|
|
2I0H
 
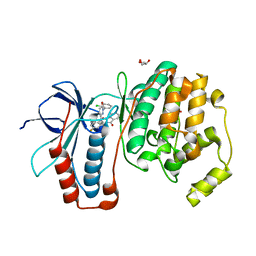 | | The structure of p38alpha in complex with an arylpyridazinone | | 分子名称: | 2-(3-{(2-CHLORO-4-FLUOROPHENYL)[1-(2-CHLOROPHENYL)-6-OXO-1,6-DIHYDROPYRIDAZIN-3-YL]AMINO}PROPYL)-1H-ISOINDOLE-1,3(2H)-DIONE, GLYCEROL, Mitogen-activated protein kinase 14 | | 著者 | Natarajan, S.R, Heller, S.T, Nam, K, Singh, S.B, Scapin, G, Patel, S, Thompson, J.E, Fitzgerald, C.E, O'Keefe, S.J. | | 登録日 | 2006-08-10 | | 公開日 | 2006-10-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | p38 MAP Kinase Inhibitors Part 6: 2-Arylpyridazin-3-ones as templates for inhibitor design.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
7NTF
 
 | |
7SU3
 
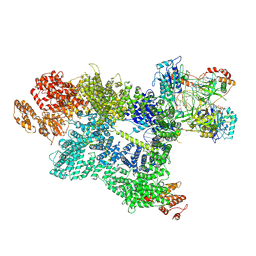 | | CryoEM structure of DNA-PK complex VII | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), ... | | 著者 | Chen, X, Liu, L, Gellert, M, Yang, W. | | 登録日 | 2021-11-16 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
7SUD
 
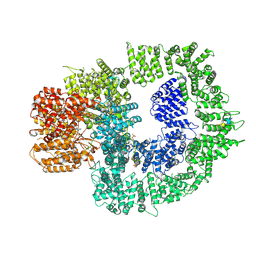 | | CryoEM structure of DNA-PK complex VIII | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, DNA-dependent protein kinase catalytic subunit, MAGNESIUM ION, ... | | 著者 | Chen, X, Liu, L, Gellert, M, Yang, W. | | 登録日 | 2021-11-16 | | 公開日 | 2022-01-12 | | 最終更新日 | 2022-01-19 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
8W9D
 
 | |
2VUS
 
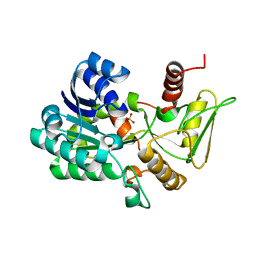 | | Crystal structure of unliganded NmrA-AreA zinc finger complex | | 分子名称: | CHLORIDE ION, NITROGEN METABOLITE REPRESSION REGULATOR NMRA, NITROGEN REGULATORY PROTEIN AREA, ... | | 著者 | Kotaka, M, Johnson, C, Lamb, H.K, Hawkins, A.R, Ren, J, Stammers, D.K. | | 登録日 | 2008-05-30 | | 公開日 | 2008-07-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural Analysis of the Recognition of the Negative Regulator Nmra and DNA by the Zinc Finger from the Gata-Type Transcription Factor Area.
J.Mol.Biol., 381, 2008
|
|
2W99
 
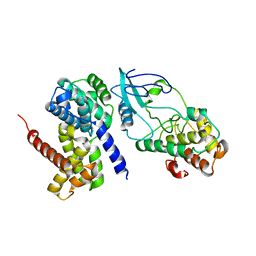 | | Crystal Structure of CDK4 in complex with a D-type cyclin | | 分子名称: | CELL DIVISION PROTEIN KINASE 4, G1/S-SPECIFIC CYCLIN-D1 | | 著者 | Day, P.J, Cleasby, A, Tickle, I.J, Reilly, M.O, Coyle, J.E, Holding, F.P, McMenamin, R.L, Yon, J, Chopra, R, Lengauer, C, Jhoti, H. | | 登録日 | 2009-01-22 | | 公開日 | 2009-03-10 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of Human Cdk4 in Complex with a D-Type Cyclin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2VUT
 
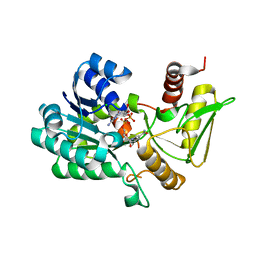 | | Crystal structure of NAD-bound NmrA-AreA zinc finger complex | | 分子名称: | CHLORIDE ION, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Kotaka, M, Johnson, C, Lamb, H.K, Hawkins, A.R, Ren, J, Stammers, D.K. | | 登録日 | 2008-05-30 | | 公開日 | 2008-07-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Analysis of the Recognition of the Negative Regulator Nmra and DNA by the Zinc Finger from the Gata-Type Transcription Factor Area.
J.Mol.Biol., 381, 2008
|
|
2VT5
 
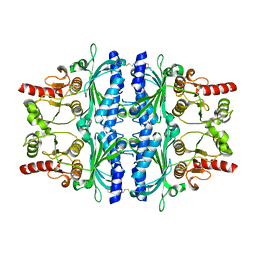 | | FRUCTOSE-1,6-BISPHOSPHATASE(D-FRUCTOSE-1,6-BISPHOSPHATE -1- PHOSPHOHYDROLASE) (E.C.3.1.3.11) COMPLEXED WITH A DUAL BINDING AMP SITE INHIBITOR | | 分子名称: | 4-AMINO-N-[(2-SULFANYLETHYL)CARBAMOYL]BENZENESULFONAMIDE, FRUCTOSE-1,6-BISPHOSPHATASE 1 | | 著者 | Ruf, A, Joseph, C, Benz, J, Fol, B, Tetaz, T. | | 登録日 | 2008-05-09 | | 公開日 | 2008-07-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Allosteric Fbpase Inhibitors Gain 10(5) Times in Potency When Simultaneously Binding Two Neighboring AMP Sites.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
8W9C
 
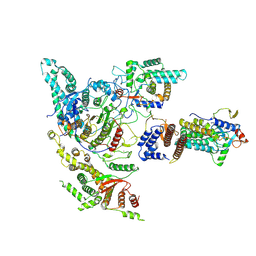 | |
6CBI
 
 | | PCNA in complex with inhibitor | | 分子名称: | GLY-ARG-LYS-ARG-ARG-GLN-DAB-SER-MET-THR-GLU-PHE-TYR-HIS, Proliferating cell nuclear antigen, SULFATE ION | | 著者 | Bruning, J.B, Wegener, K.L. | | 登録日 | 2018-02-03 | | 公開日 | 2018-07-04 | | 最終更新日 | 2020-01-29 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Rational Design of a 310-Helical PIP-Box Mimetic Targeting PCNA, the Human Sliding Clamp.
Chemistry, 24, 2018
|
|
5CQG
 
 | | Structure of Tribolium telomerase in complex with the highly specific inhibitor BIBR1532 | | 分子名称: | 2-{[(2E)-3-(naphthalen-2-yl)but-2-enoyl]amino}benzoic acid, Telomerase reverse transcriptase | | 著者 | Bryan, C, Rice, C, Hoffman, H, Harkisheimer, M, Sweeney, M, Skordalakes, E. | | 登録日 | 2015-07-21 | | 公開日 | 2015-09-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Basis of Telomerase Inhibition by the Highly Specific BIBR1532.
Structure, 23, 2015
|
|
7SJX
 
 | | Cryo-EM Structure of the PR-RT components of the HIV-1 Pol Polyprotein | | 分子名称: | Gag-Pol polyprotein | | 著者 | Lyumkis, D, Passos, D, Arnold, E, Harrison, J.J.E.K, Ruiz, F.X. | | 登録日 | 2021-10-19 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (8.2 Å) | | 主引用文献 | Cryo-EM structure of the HIV-1 Pol polyprotein provides insights into virion maturation.
Sci Adv, 8, 2022
|
|
