7BQE
 
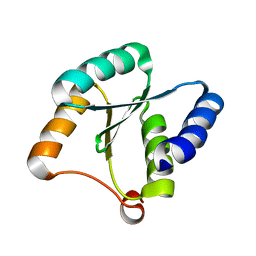 | | Solution NMR structure of NF3; de novo designed protein with a novel fold | | 分子名称: | NF3 | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | 登録日 | 2020-03-24 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQB
 
 | | Solution NMR structure of NF6; de novo designed protein with a novel fold | | 分子名称: | NF6 | | 著者 | Kobayashi, N, Nagashima, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | 登録日 | 2020-03-24 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQC
 
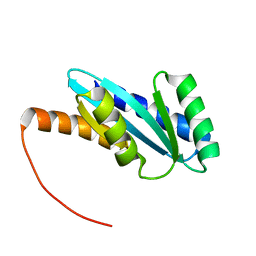 | | Solution NMR structure of NF4; de novo designed protein with a novel fold | | 分子名称: | NF4 | | 著者 | Kobayashi, N, Nagashima, T, Minami, S, Koga, R, Chikenji, T, Koga, N. | | 登録日 | 2020-03-24 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
8B16
 
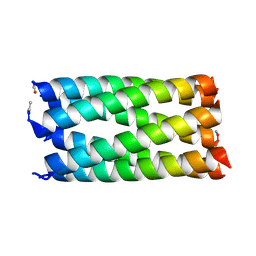 | |
8B15
 
 | |
6CO2
 
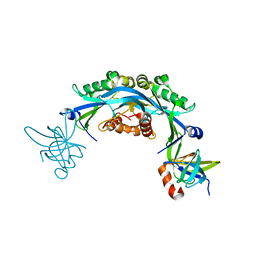 | | Structure of an engineered protein (NUDT16TI) in complex with 53BP1 Tudor domains | | 分子名称: | NUDT16-Tudor-interacting (NUDT16TI), TP53-binding protein 1 | | 著者 | Botuyan, M.V, Thompson, J.R, Cui, G, Mer, G. | | 登録日 | 2018-03-10 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4ZV6
 
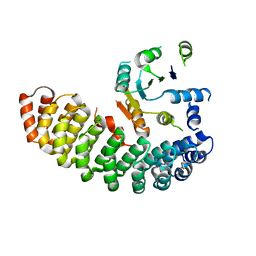 | | Crystal structure of the artificial alpharep-7 octarellinV.1 complex | | 分子名称: | AlphaRep-7, Octarellin V.1 | | 著者 | Figueroa, M, Sleutel, M, Urvoas, A, Valerio-Lepiniec, M, Minard, P, Martial, J.A, van de Weerdt, C. | | 登録日 | 2015-05-18 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | The unexpected structure of the designed protein Octarellin V.1 forms a challenge for protein structure prediction tools.
J.Struct.Biol., 195, 2016
|
|
6CO1
 
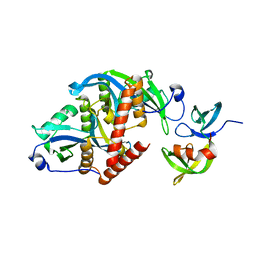 | |
6D0L
 
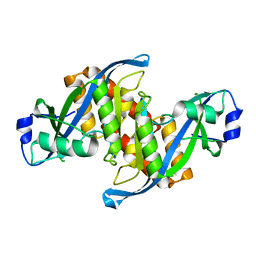 | | Structure of human TIRR | | 分子名称: | Tudor-interacting repair regulator protein | | 著者 | Cui, G, Botuyan, M.V, Mer, G. | | 登録日 | 2018-04-10 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7A4Y
 
 | |
7A4T
 
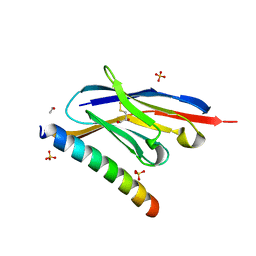 | |
7A48
 
 | |
7A4D
 
 | |
7A50
 
 | |
3Q9U
 
 | | In silico and in vitro co-evolution of a high affinity complementary protein-protein interface | | 分子名称: | COENZYME A, CoA binding protein, consensus ankyrin repeat | | 著者 | Karanicolas, J, Corn, J.E, Chen, I, Joachimiak, L.A, Dym, O, Chung, S, Albeck, S, Unger, T, Hu, W, Liu, G, Delbecq, S, Montelione, G.T, Spiegel, C, Liu, D, Baker, D, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-01-10 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A de novo protein binding pair by computational design and directed evolution.
Mol.Cell, 42, 2011
|
|
3Q9N
 
 | | In silico and in vitro co-evolution of a high affinity complementary protein-protein interface | | 分子名称: | CARBAMOYL SARCOSINE, COENZYME A, CoA binding protein, ... | | 著者 | Karanicolas, J, Corn, J.E, Chen, I, Joachimiak, L.A, Dym, O, Chung, S, Albeck, S, Unger, T, Hu, W, Liu, G, Delbecq, S, Montelione, G.T, Spiegel, C, Liu, D, Baker, D, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-01-09 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A de novo protein binding pair by computational design and directed evolution.
Mol.Cell, 42, 2011
|
|
5AYZ
 
 | | CRYSTAL STRUCTURE OF HUMAN QUINOLINATE PHOSPHORIBOSYLTRANSFERASE IN COMPLEX WITH THE PRODUCT NICOTINATE MONONUCLEOTIDE | | 分子名称: | NICOTINATE MONONUCLEOTIDE, Nicotinate-nucleotide pyrophosphorylase [carboxylating] | | 著者 | Youn, H.S, Kim, T.G, Kim, M.K, Kang, G.B, Kang, J.Y, Seo, Y.J, Lee, J.G, An, J.Y, Park, K.R, Lee, Y, Im, Y.J, Lee, J.H, Fukuoka, S.I, Eom, S.H. | | 登録日 | 2015-09-14 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
3QA9
 
 | |
2MN4
 
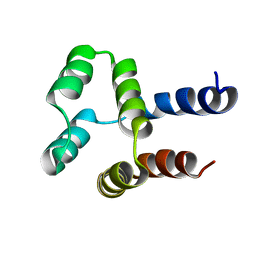 | | NMR solution structure of a computational designed protein based on structure template 1cy5 | | 分子名称: | Computational designed protein based on structure template 1cy5 | | 著者 | Xiong, P, Wang, M, Zhang, J, Chen, Q, Liu, H. | | 登録日 | 2014-03-28 | | 公開日 | 2014-10-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Protein design with a comprehensive statistical energy function and boosted by experimental selection for foldability
Nat Commun, 5, 2014
|
|
4F2V
 
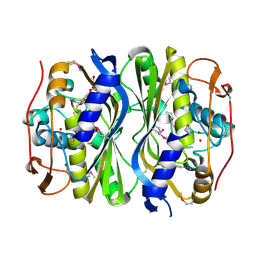 | | Crystal Structure of de novo designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR165 | | 分子名称: | DI(HYDROXYETHYL)ETHER, DODECYL-ALPHA-D-MALTOSIDE, De novo designed serine hydrolase | | 著者 | Kuzin, A, Lew, S, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Rajagopalan, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-05-08 | | 公開日 | 2012-05-30 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.493 Å) | | 主引用文献 | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
6L63
 
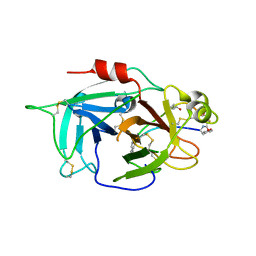 | | Human Coagulation Factor XIIa (FXIIa) bound with the macrocyclic peptide F3 containing two (1S,2S)-2-ACHC residues | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL GROUP, ... | | 著者 | Sengoku, T, Katoh, T, Hirata, K, Suga, H, Ogata, K. | | 登録日 | 2019-10-26 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Ribosomal synthesis and de novo discovery of bioactive foldamer peptides containing cyclic beta-amino acids.
Nat.Chem., 12, 2020
|
|
3O6Y
 
 | | Robust computational design, optimization, and structural characterization of retroaldol enzymes | | 分子名称: | Retro-Aldolase, SULFATE ION | | 著者 | Althoff, E.A, Wang, L, Jiang, L, Moody, J, Bolduc, J, Lassila, J.K, Wang, Z.Z, Smith, M, Hari, S, Herschlag, D, Stoddard, B.L, Baker, D. | | 登録日 | 2010-07-29 | | 公開日 | 2011-06-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.091 Å) | | 主引用文献 | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
3NXF
 
 | | Robust computational design, optimization, and structural characterization of retroaldol enzymes | | 分子名称: | Retro-Aldolase, SULFATE ION | | 著者 | Althoff, E.A, Jiang, L, Wang, L, Lassila, J.K, Moody, J, Bolduc, J, Wang, Z.Z, Smith, M, Hari, S, Herschlag, D, Stoddard, B.L, Baker, D. | | 登録日 | 2010-07-13 | | 公開日 | 2011-06-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
4DRT
 
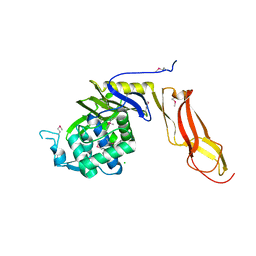 | | Three dimensional structure of de novo designed serine hydrolase OSH26, Northeast Structural Genomics Consortium (NESG) target OR89 | | 分子名称: | CHLORIDE ION, SODIUM ION, de novo designed serine hydrolase, ... | | 著者 | Kuzin, A, Su, M, Rajagopalan, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-02-17 | | 公開日 | 2012-04-18 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.002 Å) | | 主引用文献 | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4ZN8
 
 | |
