6SKQ
 
 | |
6U58
 
 | | Toho1 Beta Lactamase Glu166Gln Mutant | | 分子名称: | Beta-lactamase, SULFATE ION | | 著者 | Langan, P.S, Sullivan, B, Weiss, K.L. | | 登録日 | 2019-08-27 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | NEUTRON DIFFRACTION (1.9 Å), X-RAY DIFFRACTION | | 主引用文献 | Probing the Role of the Conserved Residue Glu166 in a Class A Beta-Lactamase Using Neutron and X-ray Protein Crystallography
Acta Crystallogr.,Sect.D, 76, 2020
|
|
6UE2
 
 | | 1.85 Angstrom Resolution Crystal Structure of Class D beta-lactamase from Clostridium difficile 630 | | 分子名称: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-09-20 | | 公開日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | 1.85 Angstrom Resolution Crystal Structure of Class D beta-lactamase from Clostridium difficile 630.
To Be Published
|
|
6SYN
 
 | | Crystal structure of Y. pestis penicillin-binding protein 3 | | 分子名称: | (2R,4S)-2-[(1R)-1-{[(2S)-2-carboxy-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, ACETATE ION, Peptidoglycan D,D-transpeptidase FtsI | | 著者 | Pankov, G, Hunter, W.N, Dawson, A. | | 登録日 | 2019-09-30 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | The structure of penicillin-binding protein 3 from Yersinia pestis
To Be Published
|
|
6T1H
 
 | | OXA-51-like beta-lactamase OXA-66 | | 分子名称: | Beta-lactamase OXA-66, ZINC ION | | 著者 | Takebayashi, Y, Chirgadze, D, Henderson, S, Warburton, P.J, Evans, B.E. | | 登録日 | 2019-10-04 | | 公開日 | 2020-10-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of the OXA-51-like beta-lactamase OXA-66
To Be Published
|
|
6UN3
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 in complex with ticarcillin | | 分子名称: | (2R,4S)-2-[(1R)-1-{[(2R)-2-carboxy-2-(thiophen-3-yl)acetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CALCIUM ION, GLYCEROL, ... | | 著者 | Sacco, M, Chen, Y. | | 登録日 | 2019-10-10 | | 公開日 | 2019-10-30 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Influence of the alpha-Methoxy Group on the Reaction of Temocillin with Pseudomonas aeruginosa PBP3 and CTX-M-14 beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2019
|
|
6UN1
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 in complex with temocillin | | 分子名称: | (2R,4S)-2-[(1S)-1-{[(2R)-2-carboxy-2-(thiophen-3-yl)acetyl]amino}-1-methoxy-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4 -carboxylic acid, Peptidoglycan D,D-transpeptidase FtsI | | 著者 | Sacco, M, Chen, Y. | | 登録日 | 2019-10-10 | | 公開日 | 2019-10-30 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Influence of the alpha-Methoxy Group on the Reaction of Temocillin with Pseudomonas aeruginosa PBP3 and CTX-M-14 beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2019
|
|
6UVK
 
 | | OXA-48 bound by inhibitor CDD-97 | | 分子名称: | 1,2-ETHANEDIOL, 1-{4-[4-(2-ethoxyphenyl)piperazin-1-yl]-1,3,5-triazin-2-yl}piperidine-4-carboxylic acid, Beta-lactamase, ... | | 著者 | Taylor, D.M, Hu, L, Prasad, B.V.V, Sankaran, B, Palzkill, T.G. | | 登録日 | 2019-11-02 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Identifying Oxacillinase-48 Carbapenemase Inhibitors Using DNA-Encoded Chemical Libraries.
Acs Infect Dis., 6, 2020
|
|
6V1O
 
 | | Structure of OXA-48 bound to QPX7728 at 1.80 A | | 分子名称: | (1aR,7bS)-5-fluoro-2-hydroxy-1,1a,2,7b-tetrahydrocyclopropa[c][1,2]benzoxaborinine-4-carboxylic acid, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Pemberton, O.A, Chen, Y. | | 登録日 | 2019-11-20 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Discovery of Cyclic Boronic Acid QPX7728, an Ultrabroad-Spectrum Inhibitor of Serine and Metallo-beta-lactamases.
J.Med.Chem., 63, 2020
|
|
6TII
 
 | |
6TIX
 
 | |
6V6N
 
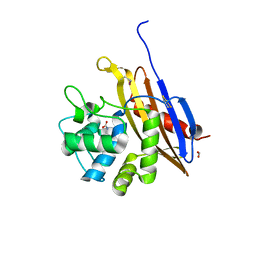 | | The crystal structure of a class D beta-lactamase from Agrobacterium tumefaciens | | 分子名称: | Beta-lactamase, FORMIC ACID, GLYCEROL, ... | | 著者 | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-12-05 | | 公開日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The crystal structure of a class D beta-lactamase from Agrobacterium tumefaciens
To Be Published
|
|
6VBC
 
 | |
6VBD
 
 | |
6VBM
 
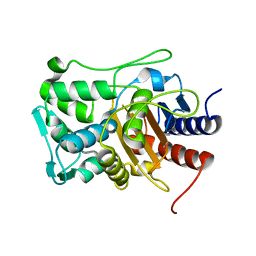 | | Crystal structure of a S310A mutant of PBP2 from Neisseria gonorrhoeae | | 分子名称: | PHOSPHATE ION, Probable peptidoglycan D,D-transpeptidase PenA | | 著者 | Singh, A, Davies, C. | | 登録日 | 2019-12-19 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Mutations in Neisseria gonorrhoeae penicillin-binding protein 2 associated with extended-spectrum cephalosporin resistance create an energetic barrier against acylation via restriction of protein dynamics
J.Biol.Chem., 2020
|
|
6VBL
 
 | |
6TUD
 
 | |
6VJE
 
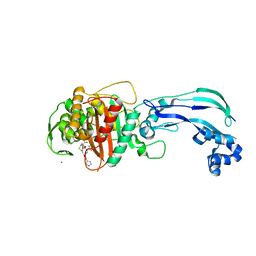 | | Crystal structure of Pseudomonas aeruginosa penicillin-binding protein 3 (PBP3) complexed with ceftobiprole | | 分子名称: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, CHLORIDE ION, Peptidoglycan D,D-transpeptidase FtsI | | 著者 | van den Akker, F, Kumar, V. | | 登録日 | 2020-01-15 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structural Insights into Ceftobiprole Inhibition of Pseudomonas aeruginosa Penicillin-Binding Protein 3.
Antimicrob.Agents Chemother., 64, 2020
|
|
6XV5
 
 | |
6VOT
 
 | | Crystal structure of Pseudomonas aerugonisa PBP3 complexed to gamma-lactam YU253434 | | 分子名称: | 1-[(2S)-2-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-3-oxopropyl]-4-{[2-(5,6 -dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl]carbamoyl}-2,5-dihydro-1H-pyrazole-3-carboxylic acid, Peptidoglycan D,D-transpeptidase FtsI | | 著者 | van den Akker, F. | | 登録日 | 2020-01-31 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A gamma-Lactam Siderophore Antibiotic Effective against Multidrug-Resistant Gram-Negative Bacilli.
J.Med.Chem., 63, 2020
|
|
6Y6Z
 
 | | Structure of Pseudomonas aeruginosa Penicillin-Binding Protein 3 (PBP3) in complex with Compound 1 | | 分子名称: | GLYCEROL, Peptidoglycan D,D-transpeptidase FtsI, ~{tert}-butyl ~{N}-[(2~{S})-2-methyl-4-oxidanyl-1-oxidanylidene-pent-4-en-2-yl]carbamate | | 著者 | Newman, H, Bellini, D, Dowson, C.G. | | 登録日 | 2020-02-27 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion.
Chem Sci, 11, 2020
|
|
6Y6U
 
 | | Structure of Pseudomonas aeruginosa Penicillin-Binding Protein 3 (PBP3) in complex with Compound 6 | | 分子名称: | 2-(4-hydroxyphenyl)-~{N}-[(2~{S})-2-methyl-4-oxidanyl-1-oxidanylidene-pent-4-en-2-yl]ethanamide, GLYCEROL, Peptidoglycan D,D-transpeptidase FtsI | | 著者 | Newman, H, Bellini, D, Dowson, C.G. | | 登録日 | 2020-02-27 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion
Chem Sci, 11, 2020
|
|
6W5G
 
 | | Class D beta-lactamase BAT-2 | | 分子名称: | 1,2-ETHANEDIOL, BAT-2 beta-lactamase | | 著者 | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | 登録日 | 2020-03-13 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.451 Å) | | 主引用文献 | A surface loop modulates activity of the Bacillus class D beta-lactamases.
J.Struct.Biol., 211, 2020
|
|
6W5E
 
 | | Class D beta-lactamase BSU-2 | | 分子名称: | 1,2-ETHANEDIOL, BSU-2 beta-lactamase, MALONATE ION | | 著者 | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | 登録日 | 2020-03-13 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | A surface loop modulates activity of the Bacillus class D beta-lactamases.
J.Struct.Biol., 211, 2020
|
|
6W5O
 
 | | Class D beta-lactamase BAT-2 delta mutant | | 分子名称: | 1,2-ETHANEDIOL, BAT-2 Beta-lactamase delta mutant, CITRATE ANION | | 著者 | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | 登録日 | 2020-03-13 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | A surface loop modulates activity of the Bacillus class D beta-lactamases.
J.Struct.Biol., 211, 2020
|
|
