3HPI
 
 | |
4ZVR
 
 | |
4ZVT
 
 | |
4ZVP
 
 | |
4ZVU
 
 | |
4ZVQ
 
 | |
4ZVS
 
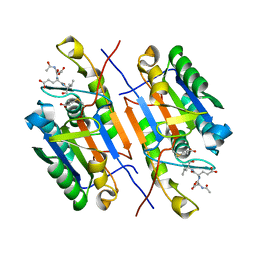 | |
4ZVO
 
 | |
8EUN
 
 | | MicroED structure of an Aeropyrum pernix protoglobin metallo-carbene complex | | 分子名称: | Protogloblin ApPgb, benzyl[3,3'-(7,12-diethenyl-3,8,13,17-tetramethylporphyrin-2,18-diyl-kappa~4~N~21~,N~22~,N~23~,N~24~)di(propanoato)(2-)]iron | | 著者 | Danelius, E, Gonen, T, Unge, J.T. | | 登録日 | 2022-10-19 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | 主引用文献 | MicroED Structure of a Protoglobin Reactive Carbene Intermediate.
J.Am.Chem.Soc., 145, 2023
|
|
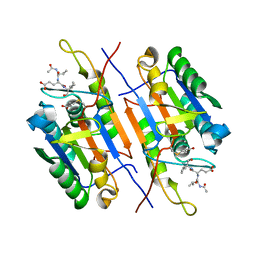
8EUM
 
 | | MicroED structure of an Aeropyrum pernix protoglobin mutant | | 分子名称: | FE (III) ION, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Danelius, E, Gonen, T, Unge, J.T. | | 登録日 | 2022-10-18 | | 公開日 | 2023-11-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (2.1 Å) | | 主引用文献 | MicroED Structure of a Protoglobin Reactive Carbene Intermediate.
J.Am.Chem.Soc., 145, 2023
|
|
4BS0
 
 | | Crystal Structure of Kemp Eliminase HG3.17 E47N,N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | 分子名称: | 6-NITROBENZOTRIAZOLE, KEMP ELIMINASE HG3.17, SULFATE ION | | 著者 | Blomberg, R, Kries, H, Pinkas, D.M, Mittl, P.R.E, Gruetter, M.G, Privett, H.K, Mayo, S, Hilvert, D. | | 登録日 | 2013-06-06 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.09 Å) | | 主引用文献 | Precision is Essential for Efficient Catalysis in an Evolved Kemp Eliminase
Nature, 503, 2013
|
|
1PVQ
 
 | | BASIS FOR A SWITCH IN SUBSTRATE SPECIFICITY: CRYSTAL STRUCTURE OF SELECTED VARIANT OF CRE SITE-SPECIFIC RECOMBINASE, LNSGG BOUND TO THE ENGINEERED RECOGNITION SITE LOXM7 | | 分子名称: | DNA 34-MER, Recombinase cre | | 著者 | Baldwin, E.P, Martin, S.S, Abel, J, Gelato, K.A, Kim, H, Schultz, P.G, Santoro, S.W. | | 登録日 | 2003-06-28 | | 公開日 | 2004-02-17 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | A specificity switch in selected cre recombinase variants is mediated by macromolecular plasticity and water.
Chem.Biol., 10, 2003
|
|
1PVR
 
 | | BASIS FOR A SWITCH IN SUBSTRATE SPECIFICITY: CRYSTAL STRUCTURE OF SELECTED VARIANT OF CRE SITE-SPECIFIC RECOMBINASE, LNSGG BOUND TO THE LOXP (WILDTYPE) RECOGNITION SITE | | 分子名称: | 34-MER, Recombinase CRE | | 著者 | Baldwin, E.P, Martin, S.S, Abel, J, Gelato, K.A, Kim, H, Schultz, P.G, Santoro, S.W. | | 登録日 | 2003-06-28 | | 公開日 | 2004-02-17 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | A specificity switch in selected cre recombinase variants is mediated by macromolecular plasticity and water.
Chem.Biol., 10, 2003
|
|
1PVP
 
 | | BASIS FOR A SWITCH IN SUBSTRATE SPECIFICITY: CRYSTAL STRUCTURE OF SELECTED VARIANT OF CRE SITE-SPECIFIC RECOMBINASE, ALSHG BOUND TO THE ENGINEERED RECOGNITION SITE LOXM7 | | 分子名称: | 34-MER, Recombinase cre | | 著者 | Baldwin, E.P, Martin, S.S, Abel, J, Gelato, K.A, Kim, H, Schultz, P.G, Santoro, S.W. | | 登録日 | 2003-06-28 | | 公開日 | 2004-02-17 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | A specificity switch in selected cre recombinase variants is mediated by macromolecular plasticity and water.
Chem.Biol., 10, 2003
|
|
6TWW
 
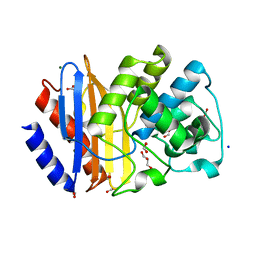 | | Variant W229D/F290W-19 of the last common ancestor of Gram-negative bacteria beta-lactamase class A (GNCA4) | | 分子名称: | ACETATE ION, Beta-Lactamase (GNCA4), FORMIC ACID, ... | | 著者 | Gavira, J.A, Risso, V, Sanchez-Ruiz, J.M, Romero-Rivera, A, Kamerlin, S.C.L. | | 登録日 | 2020-01-13 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Enhancing ade novoenzyme activity by computationally-focused ultra-low-throughput screening.
Chem Sci, 11, 2020
|
|
6TXD
 
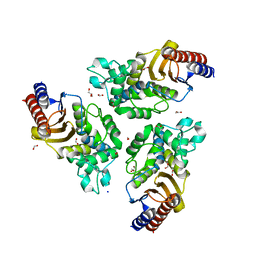 | | Variant W229D/F290W-12 of the last common ancestor of Gram-negative bacteria beta-lactamase class A (GNCA4) | | 分子名称: | ACETATE ION, Beta lactamase (GNCA4-12), FORMIC ACID, ... | | 著者 | Gavira, J.A, Risso, V, Sanchez-Ruiz, J.M, Romero-Rivera, A, Kamerlin, S.C.L. | | 登録日 | 2020-01-14 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Enhancing ade novoenzyme activity by computationally-focused ultra-low-throughput screening.
Chem Sci, 11, 2020
|
|
6TY6
 
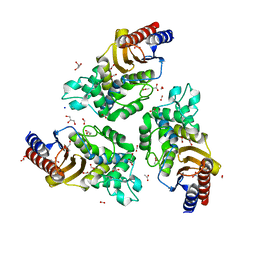 | | Variant W229D/F290W-2 of the last common ancestor of Gram-negative bacteria beta-lactamase class A (GNCA4) bound to 5(6)-nitrobenzotriazole (TS-analog) | | 分子名称: | 6-NITROBENZOTRIAZOLE, ACETATE ION, Beta lactamase (GNCA4-2), ... | | 著者 | Gavira, J.A, Risso, V, Sanchez-Ruiz, J.M, Romero-Rivera, A, Kamerlin, S.C.L, Ortega-Munoz, M, Santoyo-Gonzalez, F. | | 登録日 | 2020-01-15 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Enhancing ade novoenzyme activity by computationally-focused ultra-low-throughput screening.
Chem Sci, 11, 2020
|
|
6GWI
 
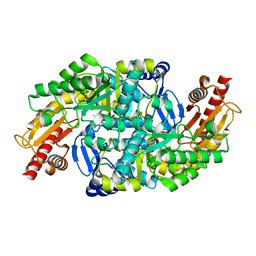 | |
8BP0
 
 | |
8BP1
 
 | |
5YAO
 
 | | The complex structure of SZ529 and expoxid | | 分子名称: | (1R,5S)-6-oxabicyclo[3.1.0]hexane, Limonene-1,2-epoxide hydrolase, SODIUM ION | | 著者 | Lian, W, Sun, Z.T, Zhou, J.H, Reetz, M.T. | | 登録日 | 2017-09-01 | | 公開日 | 2018-06-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.611 Å) | | 主引用文献 | Structural and Computational Insight into the Catalytic Mechanism of Limonene Epoxide Hydrolase Mutants in Stereoselective Transformations
J. Am. Chem. Soc., 140, 2018
|
|
5YNG
 
 | | Crystal structure of SZ348 in complex with cyclopentene oxide | | 分子名称: | (1R,5S)-6-oxabicyclo[3.1.0]hexane, Limonene-1,2-epoxide hydrolase, NICKEL (II) ION, ... | | 著者 | Wu, L, Sun, Z.T, Reetz, M.T, Zhou, J.H. | | 登録日 | 2017-10-24 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.497 Å) | | 主引用文献 | Structural and Computational Insight into the Catalytic Mechanism of Limonene Epoxide Hydrolase Mutants in Stereoselective Transformations.
J. Am. Chem. Soc., 140, 2018
|
|
5YQT
 
 | | Crystal Structure of the L74F/M78V/I80V/L114F mutant of LEH complexed with cyclopentene oxide | | 分子名称: | (1R,5S)-6-oxabicyclo[3.1.0]hexane, Limonene-1,2-epoxide hydrolase | | 著者 | Kong, X.D, Sun, Z.T, Wu, L, Reetz, M.T, Zhou, J.H, Xu, J.H. | | 登録日 | 2017-11-07 | | 公開日 | 2018-06-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and Computational Insight into the Catalytic Mechanism of Limonene Epoxide Hydrolase Mutants in Stereoselective Transformations.
J. Am. Chem. Soc., 140, 2018
|
|
3RWL
 
 | | Structure of P450pyr hydroxylase | | 分子名称: | Cytochrome P450 alkane hydroxylase 1 CYP153A7, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Pompidor, G. | | 登録日 | 2011-05-09 | | 公開日 | 2012-04-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Evolving P450pyr hydroxylase for highly enantioselective hydroxylation at non-activated carbon atom.
Chem.Commun.(Camb.), 48, 2012
|
|
7S3V
 
 | |
