7SUD
 
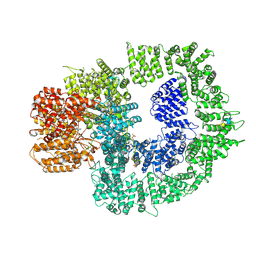 | | CryoEM structure of DNA-PK complex VIII | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, DNA-dependent protein kinase catalytic subunit, MAGNESIUM ION, ... | | 著者 | Chen, X, Liu, L, Gellert, M, Yang, W. | | 登録日 | 2021-11-16 | | 公開日 | 2022-01-12 | | 最終更新日 | 2022-01-19 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
5XWR
 
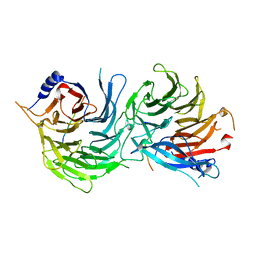 | | Crystal Structure of RBBP4-peptide complex | | 分子名称: | Histone-binding protein RBBP4, MET-SER-ARG-ARG-LYS-GLN-ALA-LYS-PRO-GLN-HIS-ILE | | 著者 | Jobichen, C, Lui, B.H, Daniel, G.T, Sivaraman, J. | | 登録日 | 2017-06-30 | | 公開日 | 2018-07-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Targeting cancer addiction for SALL4 by shifting its transcriptome with a pharmacologic peptide.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5XXQ
 
 | |
7B5L
 
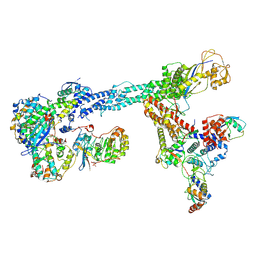 | | Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: NEDD8-CUL1-RBX1-SKP1-SKP2-CKSHS1-Cyclin A-CDK2-p27-UBE2L3~Ub~ARIH1. Transition State 1 | | 分子名称: | 5-azanylpentan-2-one, Cullin-1, Cyclin-A2, ... | | 著者 | Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | 登録日 | 2020-12-04 | | 公開日 | 2021-02-10 | | 最終更新日 | 2021-03-10 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Nature, 590, 2021
|
|
6FVU
 
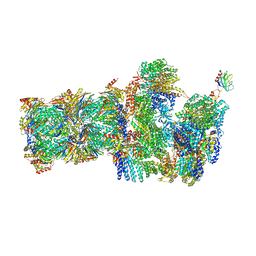 | | 26S proteasome, s2 state | | 分子名称: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | 著者 | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | 登録日 | 2018-03-05 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
6P6G
 
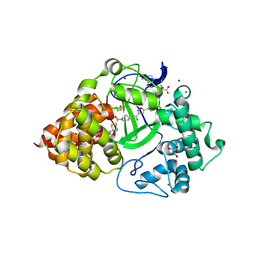 | | Co-crystal Structure of human SMYD3 with Isoxazole Amides Inhibitors | | 分子名称: | 5-cyclopropyl-N-{1-[({trans-4-[(4,4,4-trifluorobutyl)amino]cyclohexyl}methyl)sulfonyl]piperidin-4-yl}-1,2-oxazole-3-carboxamide, GLYCEROL, Histone-lysine N-methyltransferase SMYD3, ... | | 著者 | Elkins, P.A, Wang, L. | | 登録日 | 2019-06-03 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Discovery of Isoxazole Amides as Potent and Selective SMYD3 Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
6P6K
 
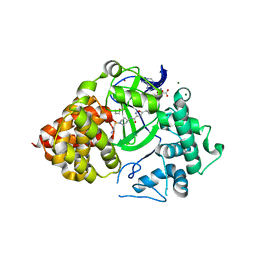 | |
7AT8
 
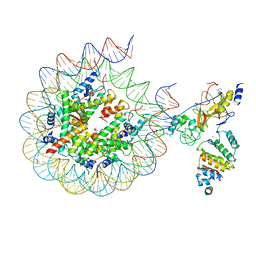 | | Histone H3 recognition by nucleosome-bound PRC2 subunit EZH2. | | 分子名称: | Histone H2A, Histone H2B 1.1, Histone H3.2, ... | | 著者 | Finogenova, K, Benda, C, Schaefer, I.B, Poepsel, S, Strauss, M, Mueller, J. | | 登録日 | 2020-10-29 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structural basis for PRC2 decoding of active histone methylation marks H3K36me2/3.
Elife, 9, 2020
|
|
8H1T
 
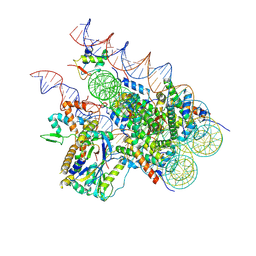 | | Cryo-EM structure of BAP1-ASXL1 bound to chromatosome | | 分子名称: | DNA (187-MER), Histone H1.4, Histone H2A type 1-D, ... | | 著者 | Ge, W, Yu, C, Xu, R.M. | | 登録日 | 2022-10-04 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Basis of the H2AK119 specificity of the Polycomb repressive deubiquitinase.
Nature, 616, 2023
|
|
5XPT
 
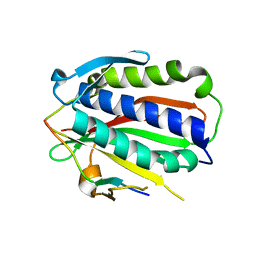 | |
5XPU
 
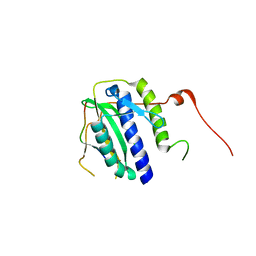 | |
8OWO
 
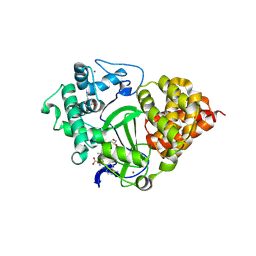 | | SMYD3 in complex with fragment FL01507 | | 分子名称: | 3-oxidanylbenzenecarbonitrile, GLYCEROL, Histone-lysine N-methyltransferase SMYD3, ... | | 著者 | Lund, B.A, Cederfelt, D, Dobritzsch, D. | | 登録日 | 2023-04-28 | | 公開日 | 2023-08-30 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Identification of fragments targeting SMYD3 using highly sensitive kinetic and multiplexed biosensor-based screening.
Rsc Med Chem, 15, 2024
|
|
1QVN
 
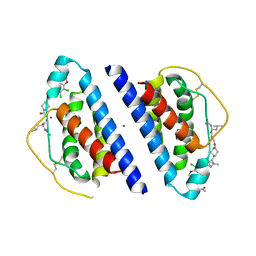 | | Structure of SP4160 Bound to IL-2 V69A | | 分子名称: | 2-GUANIDINO-4-METHYL-PENTANOIC ACID [2-(4-{5-[4-(4-ACETYLAMINO-BENZYLOXY)-2,3-DICHLORO-PHENYL]-2-METHYL-2H-PYRAZOL-3-YL}-PIPERIDIN-1-YL)-2-OXO-ETHYL]-AMIDE, Interleukin-2, ZINC ION | | 著者 | Thanos, C.D, Delano, W.L, Wells, J.A. | | 登録日 | 2003-08-28 | | 公開日 | 2005-04-05 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Hot-spot mimicry of a cytokine receptor by a small molecule.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
8H36
 
 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1-p85a dimeric complex | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H37
 
 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1-p85a tetrameric complex | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (7.52 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3F
 
 | | Cryo-EM Structure of the KBTBD2-CRL3-CSN complex | | 分子名称: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (6.73 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3A
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8(removed)-CSN complex | | 分子名称: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (7.51 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H38
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8-CSN(mutate) complex | | 分子名称: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | 著者 | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.25 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HPO
 
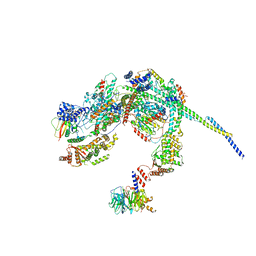 | | Cryo-EM structure of a SIN3/HDAC complex from budding yeast | | 分子名称: | Histone deacetylase RPD3, PHOSPHOTHREONINE, POTASSIUM ION, ... | | 著者 | Guo, Z, Zhan, X, Wang, C. | | 登録日 | 2022-12-12 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-07-05 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Structure of a SIN3-HDAC complex from budding yeast.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1QB3
 
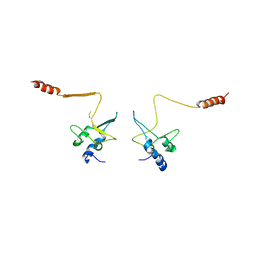 | | CRYSTAL STRUCTURE OF THE CELL CYCLE REGULATORY PROTEIN CKS1 | | 分子名称: | CYCLIN-DEPENDENT KINASES REGULATORY SUBUNIT | | 著者 | Bourne, Y, Watson, M.H, Arvai, A.S, Bernstein, S.L, Reed, S.I, Tainer, J.A. | | 登録日 | 1999-04-30 | | 公開日 | 2000-08-31 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure and mutational analysis of the Saccharomyces cerevisiae cell cycle regulatory protein Cks1: implications for domain swapping, anion binding and protein interactions.
Structure Fold.Des., 8, 2000
|
|
6FMJ
 
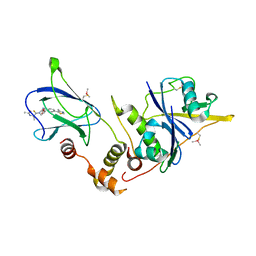 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-Acetamidopropanethioyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl) benzyl)pyrrolidine-2-carboxamide (ligand 3) | | 分子名称: | (2~{S},4~{R})-1-[(2~{S})-2-acetamidopropanethioyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | 著者 | Soares, P, Lucas, X, Ciulli, A. | | 登録日 | 2018-01-31 | | 公開日 | 2018-04-11 | | 最終更新日 | 2018-06-20 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Thioamide substitution to probe the hydroxyproline recognition of VHL ligands.
Bioorg. Med. Chem., 26, 2018
|
|
6FMK
 
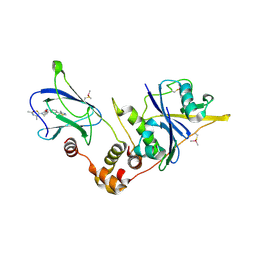 | | pVHL:EloB:EloC in complex with N-((S)-1-((2S,4R)-4-hydroxy-2-((4-(4-methylthiazol-5-yl)benzyl)carbamothioyl) pyrrolidin-1-yl)-1-thioxopropan-2-yl)acetamide (ligand 4) | | 分子名称: | Elongin-B, Elongin-C, von Hippel-Lindau disease tumor suppressor, ... | | 著者 | Soares, P, Lucas, X, Ciulli, A. | | 登録日 | 2018-01-31 | | 公開日 | 2018-04-11 | | 最終更新日 | 2018-06-20 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Thioamide substitution to probe the hydroxyproline recognition of VHL ligands.
Bioorg. Med. Chem., 26, 2018
|
|
3BWE
 
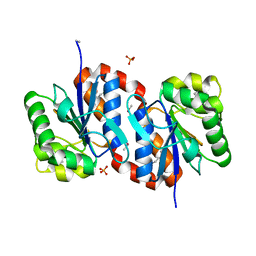 | | Crystal structure of aggregated form of DJ1 | | 分子名称: | PHOSPHATE ION, Protein DJ-1 | | 著者 | Cha, S.S. | | 登録日 | 2008-01-09 | | 公開日 | 2008-10-14 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of filamentous aggregates of human DJ-1 formed in an inorganic phosphate-dependent manner.
J.Biol.Chem., 283, 2008
|
|
7O2C
 
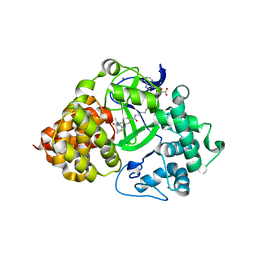 | | X-RAY STRUCTURE OF SMYD3 IN COMPLEX WITH the benzodiazepine-based probe BAY-6035 | | 分子名称: | (2S)-1-[[(1R,5S)-3-azabicyclo[3.1.0]hexan-3-yl]carbonyl]-N-(2-cyclopropylethyl)-2-methyl-4-oxidanylidene-3,5-dihydro-2H-1,5-benzodiazepine-7-carboxamide, Histone-lysine N-methyltransferase SMYD3, S-ADENOSYLMETHIONINE, ... | | 著者 | Steuber, H. | | 登録日 | 2021-03-30 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Discovery of the SMYD3 Inhibitor BAY-6035 Using Thermal Shift Assay (TSA)-Based High-Throughput Screening.
Slas Discov, 26, 2021
|
|
7O2A
 
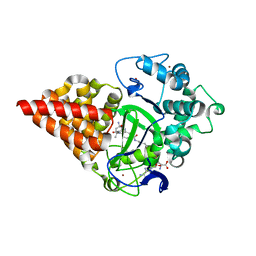 | | X-RAY STRUCTURE OF SMYD3 IN COMPLEX WITH benzodiazepine-type inhibitor compound 15 | | 分子名称: | (2S)-1-(4-azanylpiperidin-1-yl)carbonyl-N-(2-cyclopropylethyl)-2-methyl-4-oxidanylidene-3,5-dihydro-2H-1,5-benzodiazepine-7-carboxamide, Histone-lysine N-methyltransferase SMYD3, S-ADENOSYLMETHIONINE, ... | | 著者 | Steuber, H. | | 登録日 | 2021-03-30 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Discovery of the SMYD3 Inhibitor BAY-6035 Using Thermal Shift Assay (TSA)-Based High-Throughput Screening.
Slas Discov, 26, 2021
|
|
