6EN2
 
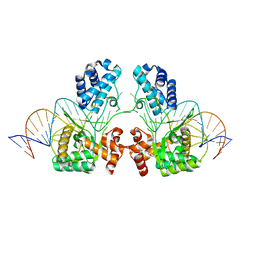 | |
6EN1
 
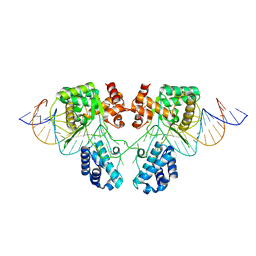 | |
7S37
 
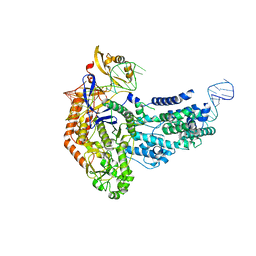 | | Cas9:sgRNA (S. pyogenes) in the open-protein conformation | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, Single-guide RNA | | 著者 | Cofsky, J.C, Soczek, K.M, Knott, G.J, Nogales, E, Doudna, J.A. | | 登録日 | 2021-09-04 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | CRISPR-Cas9 bends and twists DNA to read its sequence.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1KTQ
 
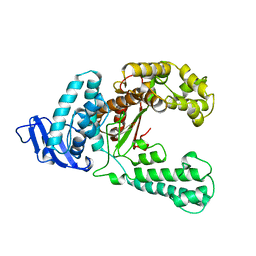 | | DNA POLYMERASE | | 分子名称: | DNA POLYMERASE I | | 著者 | Korolev, S, Waksman, G. | | 登録日 | 1995-08-16 | | 公開日 | 1996-11-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the large fragment of Thermus aquaticus DNA polymerase I at 2.5-A resolution: structural basis for thermostability.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
7DCI
 
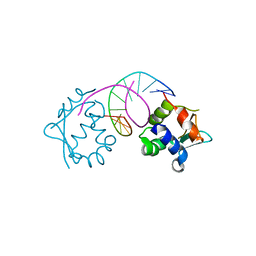 | |
8HPA
 
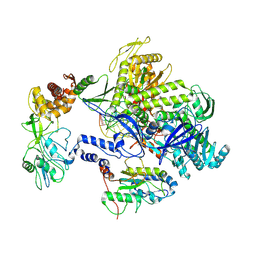 | | Monkeypox virus DNA replication holoenzyme F8, A22 and E4 complex in a DNA binding form | | 分子名称: | DNA (5'-D(*CP*GP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*TP*AP*C)-3'), DNA (5'-D(P*AP*TP*GP*GP*TP*AP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*CP*G)-3'), DNA polymerase, ... | | 著者 | Xu, Y, Wu, Y, Zhang, Y, Fan, R, Yang, Y, Li, D, Yang, B, Zhang, Z, Dong, C. | | 登録日 | 2022-12-12 | | 公開日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | Structure of DNA replication machinery from human monkeypox virus
To Be Published
|
|
5BT2
 
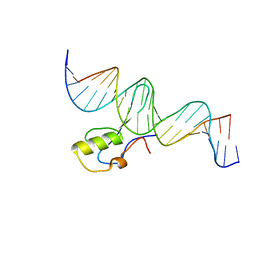 | | MeCP2 MBD domain (A140V) in complex with methylated DNA | | 分子名称: | DNA (5'-D(*AP*TP*AP*GP*AP*AP*GP*AP*AP*TP*TP*CP*(5CM)P*GP*TP*TP*CP*CP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*GP*AP*AP*(5CM)P*GP*GP*AP*AP*TP*TP*CP*TP*TP*CP*TP*A)-3'), Methyl-CpG-binding protein 2 | | 著者 | Ho, K.L, Chia, J.Y, Tan, W.S, Ng, C.L, Hu, N.J, Foo, H.L. | | 登録日 | 2015-06-02 | | 公開日 | 2016-08-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A/T Run Geometry of B-form DNA Is Independent of Bound Methyl-CpG Binding Domain, Cytosine Methylation and Flanking Sequence.
Sci Rep, 6, 2016
|
|
9C5Q
 
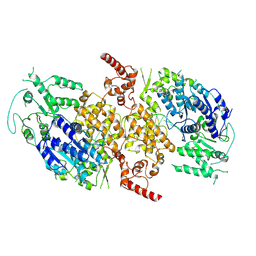 | |
6L5B
 
 | |
9ASJ
 
 | |
6L5A
 
 | |
9ASL
 
 | |
9ASK
 
 | |
5ORQ
 
 | |
6LTY
 
 | | DNA bound antitoxin HigA3 | | 分子名称: | DNA (5'-D(P*CP*CP*AP*CP*GP*AP*GP*AP*TP*AP*TP*AP*AP*CP*CP*TP*AP*GP*AP*G)-3'), DNA (5'-D(P*CP*TP*CP*TP*AP*GP*GP*TP*TP*AP*TP*AP*TP*CP*TP*CP*GP*TP*GP*G)-3'), Putative antitoxin HigA3 | | 著者 | Park, J.Y, Lee, B.J. | | 登録日 | 2020-01-23 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.28 Å) | | 主引用文献 | Induced DNA bending by unique dimerization of HigA antitoxin.
Iucrj, 7, 2020
|
|
4WKJ
 
 | | Crystallographic Structure of a Dodecameric RNA-DNA Hybrid | | 分子名称: | DNA (5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3'), MAGNESIUM ION, RNA (5'-R(*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*C)-3') | | 著者 | Davis, R.R, Shaban, N.M, Perrino, F.W, Hollis, T. | | 登録日 | 2014-10-02 | | 公開日 | 2014-10-22 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of RNA-DNA duplex provides insight into conformational changes induced by RNase H binding.
Cell Cycle, 14, 2015
|
|
9FFF
 
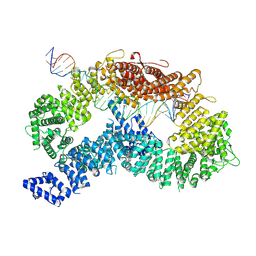 | |
9FFB
 
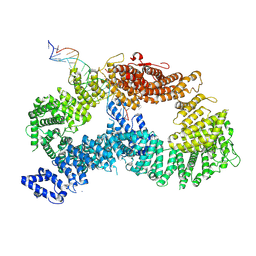 | | ss-dsDNA-FANCD2-FANCI complex | | 分子名称: | DNA (5'-D(P*CP*GP*AP*TP*GP*TP*CP*TP*CP*TP*AP*GP*AP*CP*AP*GP*CP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*AP*GP*CP*TP*GP*TP*CP*TP*AP*GP*AP*GP*AP*CP*AP*TP*CP*GP*AP*T)-3'), Fanconi anemia complementation group I, ... | | 著者 | Alcon, P, Passmore, L.A. | | 登録日 | 2024-05-22 | | 公開日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (3.59 Å) | | 主引用文献 | ss-dsDNA-FANCD2-FANCI complex
To Be Published
|
|
7D9K
 
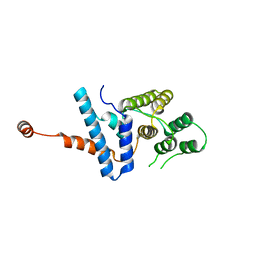 | |
7D9Y
 
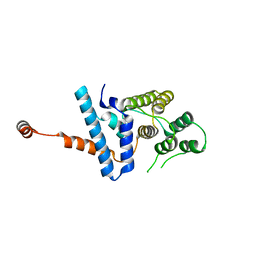 | |
6ZHE
 
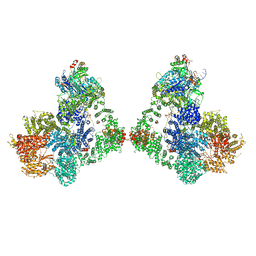 | | Cryo-EM structure of DNA-PK dimer | | 分子名称: | DNA (25-MER), DNA (26-MER), DNA (27-MER), ... | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-23 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (7.24 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZHA
 
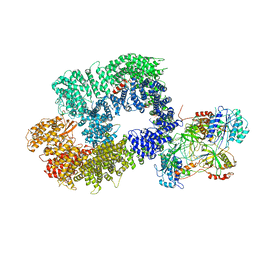 | | Cryo-EM structure of DNA-PK monomer | | 分子名称: | DNA, DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5, ... | | 著者 | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | 登録日 | 2020-06-21 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.91 Å) | | 主引用文献 | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3ZH2
 
 | | Structure of Plasmodium falciparum lactate dehydrogenase in complex with a DNA aptamer | | 分子名称: | DNA APTAMER, L-LACTATE DEHYDROGENASE | | 著者 | Cheung, Y.W, Kwok, J, Law, A.W.L, Watt, R.M, Kotaka, M, Tanner, J.A. | | 登録日 | 2012-12-20 | | 公開日 | 2013-09-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Basis for Discriminatory Recognition of Plasmodium Lactate Dehydrogenase by a DNA Aptamer
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8POP
 
 | | HK97 small terminase in complex with DNA | | 分子名称: | DNA (31-MER), Terminase small subunit | | 著者 | Chechik, M, Greive, S.J, Antson, A.A, Jenkins, H.T. | | 登録日 | 2023-07-05 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-08-21 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for DNA recognition by a viral genome-packaging machine.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4R64
 
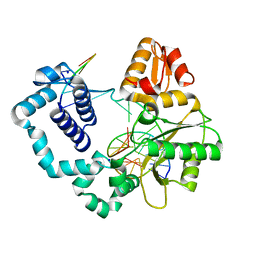 | | Binary complex crystal structure of E295K mutant of DNA polymerase Beta | | 分子名称: | DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | 著者 | Batra, V.K, Beard, W.A, Wilson, S.H. | | 登録日 | 2014-08-22 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Substrate-induced DNA Polymerase beta Activation.
J.Biol.Chem., 289, 2014
|
|
