7TNN
 
 | |
2RC7
 
 | |
7TNL
 
 | |
7TNJ
 
 | |
7TNP
 
 | |
6NJL
 
 | |
2RC8
 
 | |
2RC9
 
 | |
6NJN
 
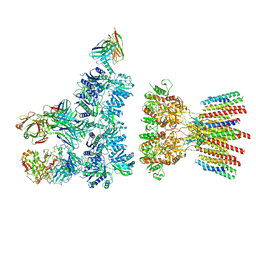 | |
4NF4
 
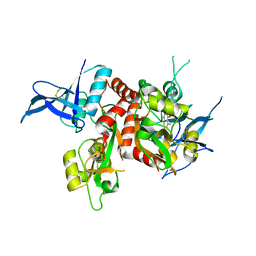 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with DCKA and glutamate | | 分子名称: | 4-hydroxy-5,7-dimethylquinoline-2-carboxylic acid, GLUTAMIC ACID, Glutamate receptor ionotropic, ... | | 著者 | Annie, J, Tajima, N, Furukawa, H. | | 登録日 | 2013-10-30 | | 公開日 | 2014-03-12 | | 最終更新日 | 2017-08-09 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Insights into Competitive Antagonism in NMDA Receptors.
Neuron, 81, 2014
|
|
2UXA
 
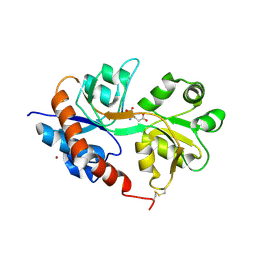 | | Crystal structure of the GluR2-flip ligand binding domain, r/g unedited. | | 分子名称: | GLUTAMATE RECEPTOR SUBUNIT GLUR2-FLIP, GLUTAMIC ACID, ZINC ION | | 著者 | Greger, I.H, Akamine, P, Khatri, L, Ziff, E.B. | | 登録日 | 2007-03-27 | | 公開日 | 2007-04-10 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Developmentally Regulated, Combinatorial RNA Processing Modulates Ampa Receptor Biogenesis.
Neuron, 51, 2006
|
|
6O9G
 
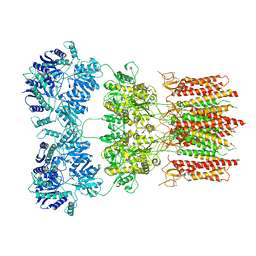 | | Open state GluA2 in complex with STZ and blocked by AgTx-636, after micelle signal subtraction | | 分子名称: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, ... | | 著者 | Twomey, E.C, Yelshanskaya, M.V, Vassilevski, A.A, Sobolevsky, A.I. | | 登録日 | 2019-03-13 | | 公開日 | 2019-03-20 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors.
Neuron, 99, 2018
|
|
4NF6
 
 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with glycine and PPDA | | 分子名称: | (2S,3R)-1-(phenanthren-2-ylcarbonyl)piperazine-2,3-dicarboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | 著者 | Jespersen, A, Tajima, N, Furukawa, H. | | 登録日 | 2013-10-30 | | 公開日 | 2014-03-12 | | 最終更新日 | 2017-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Insights into Competitive Antagonism in NMDA Receptors.
Neuron, 81, 2014
|
|
2V3U
 
 | | Structure of the ligand-binding core of the ionotropic glutamate receptor-like GluRdelta2 in complex with D-serine | | 分子名称: | CHLORIDE ION, D-SERINE, GLUTAMATE RECEPTOR DELTA-2 SUBUNIT, ... | | 著者 | Naur, P, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | 登録日 | 2007-06-22 | | 公開日 | 2007-08-07 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Ionotropic Glutamate-Like Receptor {Delta}2 Binds D-Serine and Glycine.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
6OVD
 
 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 3-ethylphenyl-ACEPC | | 分子名称: | (3S,5S)-5-[(2R)-2-amino-2-carboxyethyl]-1-(3-ethylphenyl)pyrazolidine-3-carboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | 著者 | Syrenne, J.T, Mou, T.C, Tamborini, L, Pinto, A, Sprang, S.R, Hansen, K.B. | | 登録日 | 2019-05-07 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.102 Å) | | 主引用文献 | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 3-ethylphenyl-ACEPC
To Be Published
|
|
2V3T
 
 | | Structure of the ligand-binding core of the ionotropic glutamate receptor-like GluRdelta2 in the apo form | | 分子名称: | CALCIUM ION, GLUTAMATE RECEPTOR DELTA-2 SUBUNIT SYNONYM GLURDELTA2, GLUR DELTA-2 | | 著者 | Naur, P, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | 登録日 | 2007-06-22 | | 公開日 | 2007-08-07 | | 最終更新日 | 2019-07-24 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Ionotropic Glutamate-Like Receptor {Delta}2 Binds D-Serine and Glycine.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
6ODL
 
 | | Crystal structure of GluN2A agonist binding domain with 4-butyl-(S)-CCG-IV | | 分子名称: | (1S,2R)-2-[(S)-amino(carboxy)methyl]-1-butylcyclopropane-1-carboxylic acid, Glutamate receptor ionotropic, NMDA 2A,Glutamate receptor ionotropic, ... | | 著者 | Mou, T.C, Clausen, R.P, Sprang, S.R, Hansen, K.B. | | 登録日 | 2019-03-26 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Stereoselective synthesis of novel 2'-(S)-CCG-IV analogues as potent NMDA receptor agonists.
Eur.J.Med.Chem., 212, 2021
|
|
4NF8
 
 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with glycine and glutamate in PEG2000MME | | 分子名称: | GLUTAMIC ACID, GLYCINE, Glutamate receptor ionotropic, ... | | 著者 | Jespersen, A, Tajima, N, Furukawa, H. | | 登録日 | 2013-10-30 | | 公開日 | 2014-03-12 | | 最終更新日 | 2017-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.856 Å) | | 主引用文献 | Structural Insights into Competitive Antagonism in NMDA Receptors.
Neuron, 81, 2014
|
|
6OVE
 
 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 4-propylphenyl-ACEPC | | 分子名称: | (3R,5S)-5-[(2R)-2-amino-2-carboxyethyl]-1-(4-propylphenyl)pyrazolidine-3-carboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | 著者 | Syrenne, J.T, Mou, T.C, Tamborini, L, Pinto, A, Sprang, S.R, Hansen, K.B. | | 登録日 | 2019-05-07 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 3-ethylphenyl-ACEPC
To Be Published
|
|
4O3A
 
 | | Crystal structure of the glua2 ligand-binding domain in complex with L-aspartate at 1.80 a resolution | | 分子名称: | ACETATE ION, ASPARTIC ACID, CHLORIDE ION, ... | | 著者 | Krintel, C, Frydenvang, F, Gajhede, M, Kastrup, J.S. | | 登録日 | 2013-12-18 | | 公開日 | 2014-04-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | L-Asp is a useful tool in the purification of the ionotropic glutamate receptor A2 ligand-binding domain.
Febs J., 281, 2014
|
|
4O3B
 
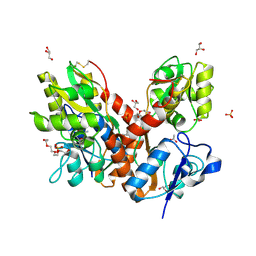 | | Crystal structure of an open/closed glua2 ligand-binding domain dimer at 1.91 A resolution | | 分子名称: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Krintel, C, de Rabassa, A.C, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | 登録日 | 2013-12-18 | | 公開日 | 2014-04-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.906 Å) | | 主引用文献 | L-Asp is a useful tool in the purification of the ionotropic glutamate receptor A2 ligand-binding domain.
Febs J., 281, 2014
|
|
4PE5
 
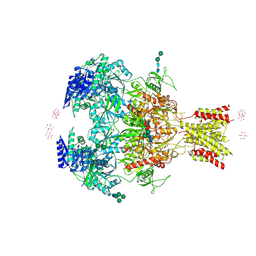 | | Crystal Structure of GluN1a/GluN2B NMDA Receptor Ion Channel | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(1R,2S)-2-(4-benzylpiperidin-1-yl)-1-hydroxypropyl]phenol, ... | | 著者 | Karakas, E, Furukawa, H. | | 登録日 | 2014-04-22 | | 公開日 | 2014-06-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.96 Å) | | 主引用文献 | Crystal structure of a heterotetrameric NMDA receptor ion channel.
Science, 344, 2014
|
|
4N07
 
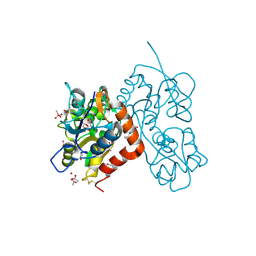 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and BPAM-344 at 1.87 A resolution | | 分子名称: | 4-cyclopropyl-7-fluoro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, CACODYLATE ION, ... | | 著者 | Noerholm, A.B, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2013-10-01 | | 公開日 | 2013-11-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Synthesis, pharmacological and structural characterization, and thermodynamic aspects of GluA2-positive allosteric modulators with a 3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide scaffold.
J.Med.Chem., 56, 2013
|
|
4NWD
 
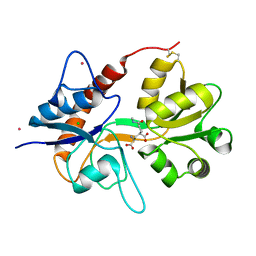 | | Crystal structure of the kainate receptor GluK3 ligand-binding domain in complex with the agonist (2S,4R)-4-(3-Methylamino-3-oxopropyl)glutamic acid at 2.6 A resolution | | 分子名称: | (4R)-4-[3-(methylamino)-3-oxopropyl]-L-glutamic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | 著者 | Venskutonyte, R, Larsen, A.P, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | 登録日 | 2013-12-06 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Molecular Recognition of Two 2,4-syn-Functionalized (S)-Glutamate Analogues by the Kainate Receptor GluK3 Ligand Binding Domain.
Chemmedchem, 9, 2014
|
|
4NWC
 
 | | Crystal structure of the GluK3 ligand-binding domain (S1S2) in complex with the agonist (2S,4R)-4-(3-Methoxy-3-oxopropyl)glutamic acid at 2.01 A resolution. | | 分子名称: | (2S,4R)-4-(3-Methoxy-3-oxopropyl) glutamic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | 著者 | Larsen, A.P, Venskutonyte, R, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | 登録日 | 2013-12-06 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.012 Å) | | 主引用文献 | Molecular Recognition of Two 2,4-syn-Functionalized (S)-Glutamate Analogues by the Kainate Receptor GluK3 Ligand Binding Domain.
Chemmedchem, 9, 2014
|
|
