4MT7
 
 | |
6MU0
 
 | |
6MVR
 
 | | Structure of a bacterial ALDH16 | | 分子名称: | Aldehyde dehydrogenase, GLYCEROL, SULFATE ION | | 著者 | Tanner, J.J, Liu, L. | | 登録日 | 2018-10-28 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J. Mol. Biol., 431, 2019
|
|
6MX2
 
 | |
4MU9
 
 | |
6MQS
 
 | |
4N14
 
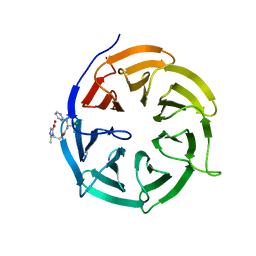 | | Crystal structure of Cdc20 and apcin complex | | 分子名称: | 2-(2-methyl-5-nitro-1H-imidazol-1-yl)ethyl [(1R)-2,2,2-trichloro-1-(pyrimidin-2-ylamino)ethyl]carbamate, Cell division cycle protein 20 homolog | | 著者 | Luo, X, Tian, W, Yu, H. | | 登録日 | 2013-10-03 | | 公開日 | 2014-08-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C.
Nature, 514, 2014
|
|
6MYN
 
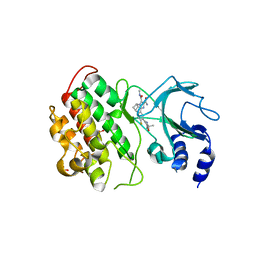 | | Crystal structure of murine NF-kappaB inducing kinase (NIK) bound to inhibitor R7 | | 分子名称: | (5s,7s)-9-fluoro-10-[(3R)-3-hydroxy-3-(5-methyl-1,2-oxazol-3-yl)but-1-yn-1-yl]-N~3~-methyl-6,7-dihydro-5H-5,7-methanoimidazo[2,1-a][2]benzazepine-2,3-dicarboxamide, Mitogen-activated protein kinase kinase kinase 14, SULFATE ION | | 著者 | Harris, S.F, Smith, M, Barker, J. | | 登録日 | 2018-11-01 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.744 Å) | | 主引用文献 | Structure Based Design of Potent Selective Inhibitors of Protein Kinase D1 (PKD1).
Acs Med.Chem.Lett., 10, 2019
|
|
6MS2
 
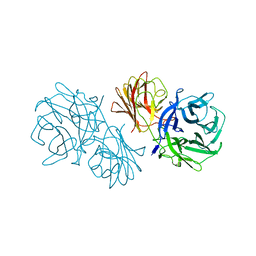 | | Crystal structure of the GH43 BlXynB protein from Bacillus licheniformis | | 分子名称: | CALCIUM ION, Glycoside Hydrolase Family 43 | | 著者 | Zanphorlin, L.M, Morais, M.A.B, Diogo, J.A, Murakami, M.T. | | 登録日 | 2018-10-16 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.494 Å) | | 主引用文献 | Structure-guided design combined with evolutionary diversity led to the discovery of the xylose-releasing exo-xylanase activity in the glycoside hydrolase family 43.
Biotechnol. Bioeng., 116, 2019
|
|
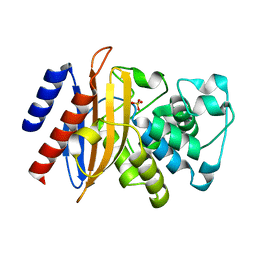
6MU9
 
 | |
4N6D
 
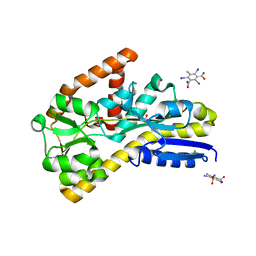 | | Crystal structure of a TRAP periplasmic solute binding protein from Desulfovibrio salexigens DSM2638 (Desal_3247), Target EFI-510112, phased with I3C, open complex, C-terminus of symmetry mate bound in ligand binding site | | 分子名称: | 1,2-ETHANEDIOL, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, CHLORIDE ION, ... | | 著者 | Vetting, M.W, Al Obaidi, N.F, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-10-11 | | 公開日 | 2013-10-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
6N4W
 
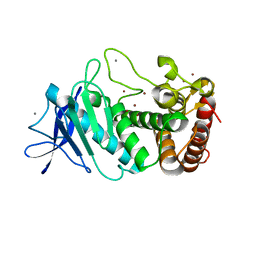 | | Tetragonal thermolysin (with 50% xylose) cryocooled in a nitrogen gas stream to 100 K | | 分子名称: | CALCIUM ION, Thermolysin, ZINC ION, ... | | 著者 | Juers, D.H, Harrison, K, Wu, B. | | 登録日 | 2018-11-20 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | A comparison of gas stream cooling and plunge cooling of macromolecular crystals.
J.Appl.Crystallogr., 52, 2019
|
|
6N56
 
 | |
6MYV
 
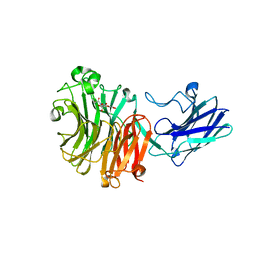 | | Sialidase26 co-crystallized with DANA-Gc | | 分子名称: | 2,6-anhydro-3,5-dideoxy-5-[(hydroxyacetyl)amino]-D-glycero-L-altro-non-2-enonic acid, Sialidase26 | | 著者 | Zaramela, L.S, Martino, C, Alisson-Silva, F, Rees, S.D, Diaz, S.L, Chuzel, L, Ganatra, M.B, Taron, C.H, Zuniga, C, Huang, J, Siegel, D, Chang, G, Varki, A, Zengler, K. | | 登録日 | 2018-11-02 | | 公開日 | 2019-10-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates.
Nat Microbiol, 4, 2019
|
|
4N7O
 
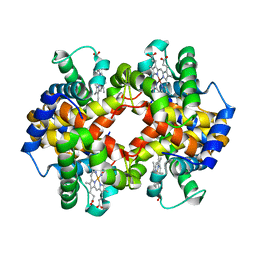 | | Capturing the haemoglobin allosteric transition in a single crystal form; Crystal structure of half-liganded human haemoglobin with phosphate at 2.5 A resolution. | | 分子名称: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Sugiyama, K, Shibayama, N, Park, S.Y. | | 登録日 | 2013-10-16 | | 公開日 | 2014-04-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.502 Å) | | 主引用文献 | Capturing the hemoglobin allosteric transition in a single crystal form
J.Am.Chem.Soc., 136, 2014
|
|
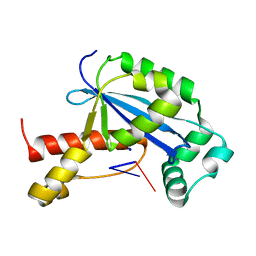
6N6E
 
 | |
6N72
 
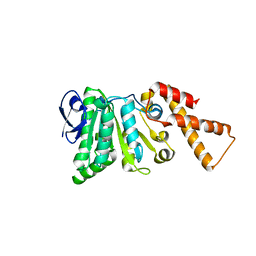 | |
4N8O
 
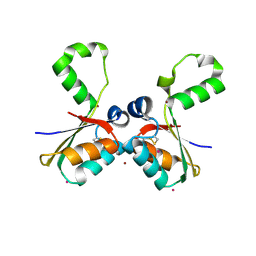 | |
4MQD
 
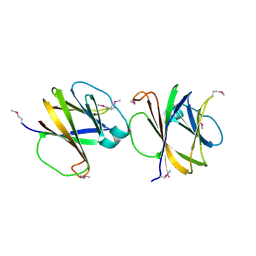 | | Crystal structure of ComJ, inhibitor of the DNA degrading activity of NucA, from Bacillus subtilis | | 分子名称: | DNA-entry nuclease inhibitor | | 著者 | Chang, C, Mack, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-09-16 | | 公開日 | 2013-10-09 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Crystal structure of ComJ, inhibitor of the DNA degrading activity of NucA, from Bacillus subtilis
To be Published
|
|
6N3K
 
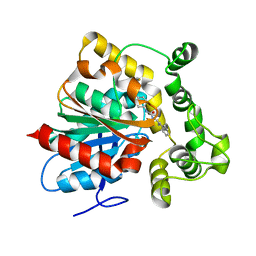 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 1 | | 分子名称: | Epoxide hydrolase, N-{cis-4-[(2,6-difluorophenyl)methoxy]cyclohexyl}-N'-(3-phenylpropyl)urea | | 著者 | de Oliveira, G.S, Adriani, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | 登録日 | 2018-11-15 | | 公開日 | 2019-11-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors.
Int. J. Biol. Macromol., 129, 2019
|
|
6N5D
 
 | |
6N5S
 
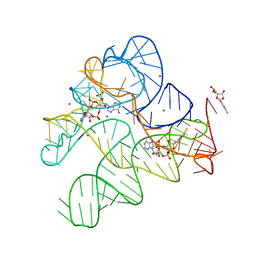 | | Structure of Human pir-miRNA-320b-2 Apical Loop and One-base-pair Stem Fused to the YdaO Riboswitch Scaffold | | 分子名称: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Shoffner, G.M, Peng, Z, Guo, F. | | 登録日 | 2018-11-22 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.802 Å) | | 主引用文献 | Three-dimensional structures of pri-miRNA apical junctions and loops revealed by scaffold-directed crystallography
To Be Published
|
|
4MT4
 
 | |
6N1I
 
 | | Cryo-EM structure of NLRC4-CARD filament | | 分子名称: | NLR family CARD domain-containing protein 4 | | 著者 | Li, Y, Fu, T, Wu, H. | | 登録日 | 2018-11-08 | | 公開日 | 2018-12-05 | | 最終更新日 | 2025-05-28 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Cryo-EM structures of ASC and NLRC4 CARD filaments reveal a unified mechanism of nucleation and activation of caspase-1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6N1Z
 
 | |
