6TD3
 
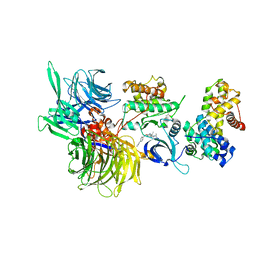 | | Structure of DDB1 bound to CR8-engaged CDK12-cyclinK | | 分子名称: | (2R)-2-({9-(1-methylethyl)-6-[(4-pyridin-2-ylbenzyl)amino]-9H-purin-2-yl}amino)butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Bunker, R.D, Petzold, G, Kozicka, Z, Thoma, N.H. | | 登録日 | 2019-11-07 | | 公開日 | 2020-06-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.46 Å) | | 主引用文献 | The CDK inhibitor CR8 acts as a molecular glue degrader that depletes cyclin K.
Nature, 585, 2020
|
|
5JK7
 
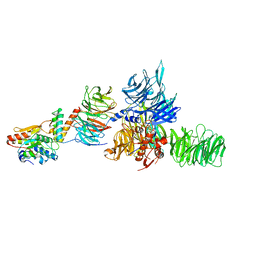 | | The X-ray structure of the DDB1-DCAF1-Vpr-UNG2 complex | | 分子名称: | DNA damage-binding protein 1, Protein VPRBP, Protein Vpr, ... | | 著者 | Calero, G, Ahn, J, Wu, Y. | | 登録日 | 2016-04-26 | | 公開日 | 2016-10-05 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.49 Å) | | 主引用文献 | The DDB1-DCAF1-Vpr-UNG2 crystal structure reveals how HIV-1 Vpr steers human UNG2 toward destruction.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4CI3
 
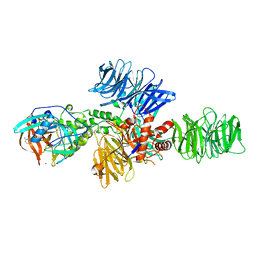 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to Pomalidomide | | 分子名称: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Pomalidomide, ... | | 著者 | Fischer, E.S, Boehm, K, Thoma, N.H. | | 登録日 | 2013-12-05 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
8BUI
 
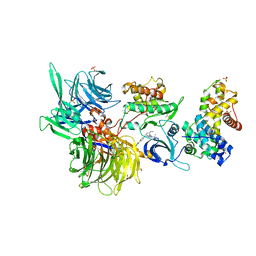 | | Structure of DDB1 bound to DRF-053-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[9-propan-2-yl-6-[(4-pyridin-2-ylphenyl)amino]purin-2-yl]amino]butan-1-ol, 1,2-ETHANEDIOL, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8B3I
 
 | | CRL4CSA-E2-Ub (state 2) | | 分子名称: | Cullin-4A, DNA damage-binding protein 1, DNA excision repair protein ERCC-8, ... | | 著者 | Kokic, G, Cramer, P. | | 登録日 | 2022-09-16 | | 公開日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | C(N)RL4CSA-E2-Ub (state 2)
To Be Published
|
|
6BN7
 
 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET23 PROTAC. | | 分子名称: | Bromodomain-containing protein 4, DNA damage-binding protein 1, Protein cereblon, ... | | 著者 | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | 登録日 | 2017-11-16 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.501 Å) | | 主引用文献 | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
8BU9
 
 | | Structure of DDB1 bound to roscovitine-engaged CDK12-cyclin K | | 分子名称: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.51 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUG
 
 | | Structure of DDB1 bound to HQ461-engaged CDK12-cyclin K | | 分子名称: | 2-[2-[(6-methylpyridin-2-yl)amino]-1,3-thiazol-4-yl]-~{N}-(5-methyl-1,3-thiazol-2-yl)ethanamide, CITRIC ACID, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.53 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
6SJ7
 
 | |
6UML
 
 | | Structural Basis for Thalidomide Teratogenicity Revealed by the Cereblon-DDB1-SALL4-Pomalidomide Complex | | 分子名称: | DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | 著者 | Clayton, T.L, Matyskiela, M.E, Pagarigan, B.E, Tran, E.T, Chamberlain, P.P. | | 登録日 | 2019-10-09 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.58 Å) | | 主引用文献 | Crystal structure of the SALL4-pomalidomide-cereblon-DDB1 complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8BUO
 
 | | Structure of DDB1 bound to DS24-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[6-[(3-fluoranyl-4-pyridin-2-yl-phenyl)methylamino]-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.58 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
5HXB
 
 | | Cereblon in complex with DDB1, CC-885, and GSPT1 | | 分子名称: | 1-(3-chloro-4-methylphenyl)-3-({2-[(3S)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-5-yl}methyl)urea, DNA damage-binding protein 1, Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, ... | | 著者 | Chamberlain, P.P, Matyskiela, M, Pagarigan, B. | | 登録日 | 2016-01-30 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | A novel cereblon modulator recruits GSPT1 to the CRL4(CRBN) ubiquitin ligase.
Nature, 535, 2016
|
|
8D80
 
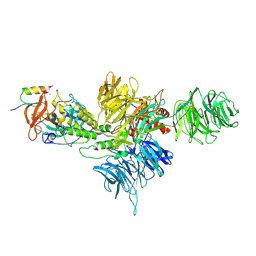 | | Cereblon~DDB1 bound to Iberdomide and Ikaros ZF1-2-3 | | 分子名称: | (3S)-3-[4-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, DNA-binding protein Ikaros, ... | | 著者 | Watson, E.R, Lander, G.C. | | 登録日 | 2022-06-07 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
4A0A
 
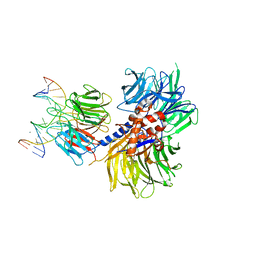 | | Structure of hsDDB1-drDDB2 bound to a 16 bp CPD-duplex (pyrimidine at D-1 position) at 3.6 A resolution (CPD 3) | | 分子名称: | 5'-D(*CP*CP*TP*GP*CP*TP*CP*CP*TP*TP*TP*CP*AP*CP*CP*C)-3', 5'-D(*GP*GP*TP*GP*AP*AP*AP*(TTD)P*AP*GP*CP*AP*GP*DGP)-3', CALCIUM ION, ... | | 著者 | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | 登録日 | 2011-09-08 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
8BUJ
 
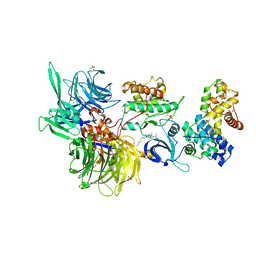 | | Structure of DDB1 bound to DS06-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[6-(octylamino)-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.62 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
6XK9
 
 | | Cereblon in complex with DDB1, CC-90009, and GSPT1 | | 分子名称: | 2-(4-chlorophenyl)-N-({2-[(3S)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-5-yl}methyl)-2,2-difluoroacetamide, DNA damage-binding protein 1, Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, ... | | 著者 | Clayton, T.L, Tran, E.T, Zhu, J, Pagarigan, B.E, Matyskiela, M.E, Chamberlain, P.P. | | 登録日 | 2020-06-25 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.64 Å) | | 主引用文献 | CC-90009, a novel cereblon E3 ligase modulator, targets acute myeloid leukemia blasts and leukemia stem cells.
Blood, 137, 2021
|
|
8BUR
 
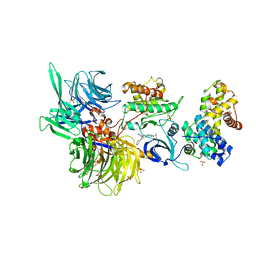 | | Structure of DDB1 bound to DS50-engaged CDK12-cyclin K | | 分子名称: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (3.64 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
7V7C
 
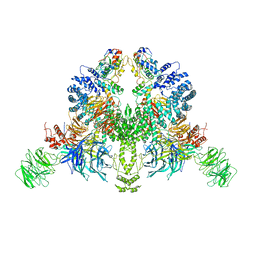 | | CryoEM structure of DDB1-VprBP-Vpr-UNG2(94-313) complex | | 分子名称: | DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, Protein Vpr, ... | | 著者 | Wang, D, Xu, J, Liu, Q, Xiang, Y. | | 登録日 | 2021-08-21 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural insights into the HIV-1 Vpr mediated ubiquitination through the Cullin-RING E3 ubiquitin ligase
To Be Published
|
|
8DEY
 
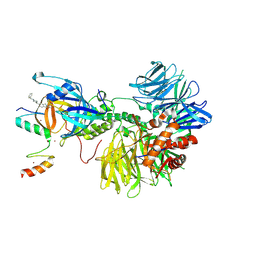 | | Ternary complex structure of Cereblon-DDB1 bound to IKZF2(ZF2,3) and the molecular glue DKY709 | | 分子名称: | (3S)-3-[5-(1-benzylpiperidin-4-yl)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, Protein cereblon, ... | | 著者 | Ma, X, Ornelas, E, Clifton, M.C. | | 登録日 | 2022-06-21 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Discovery and characterization of a selective IKZF2 glue degrader for cancer immunotherapy.
Cell Chem Biol, 30, 2023
|
|
8OV6
 
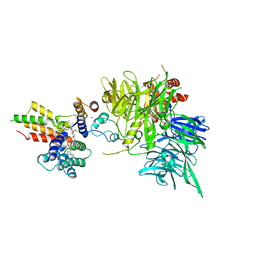 | | Ternary structure of intramolecular bivalent glue degrader IBG1 bound to BRD4 and DCAF16:DDB1deltaBPB | | 分子名称: | Bromodomain-containing protein 4, DDB1- and CUL4-associated factor 16, DDB1deltaBPB, ... | | 著者 | Cowan, A.D, Sundaramoorthy, R, Nakasone, M.A, Ciulli, A. | | 登録日 | 2023-04-25 | | 公開日 | 2023-05-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.77 Å) | | 主引用文献 | Targeted protein degradation via intramolecular bivalent glues.
Nature, 627, 2024
|
|
7ZN7
 
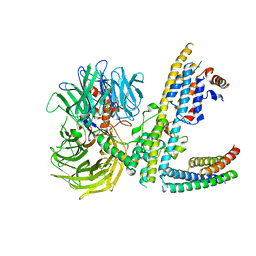 | | Cryo-EM structure of RCMV-E E27 bound to human DDB1 (deltaBPB) and rat STAT2 CCD | | 分子名称: | B27a, DNA damage-binding protein 1, Signal transducer and activator of transcription, ... | | 著者 | Lauer, S, Spahn, C.M.T, Schwefel, D. | | 登録日 | 2022-04-20 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.78 Å) | | 主引用文献 | Structural mechanism of CRL4-instructed STAT2 degradation via a novel cytomegaloviral DCAF receptor.
Embo J., 42, 2023
|
|
7LPS
 
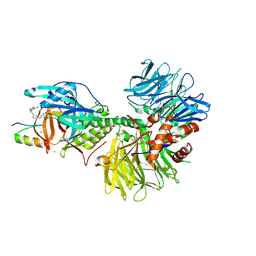 | | Crystal structure of DDB1-CRBN-ALV1 complex bound to Helios (IKZF2 ZF2) | | 分子名称: | 3-[3-[[1-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2,5-bis(oxidanylidene)pyrrol-3-yl]amino]phenyl]-~{N}-(3-chloranyl-4-methyl-phenyl)propanamide, DNA damage-binding protein 1, Protein cereblon, ... | | 著者 | Nowak, R.P, Fischer, E.S. | | 登録日 | 2021-02-12 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.78 Å) | | 主引用文献 | Acute pharmacological degradation of Helios destabilizes regulatory T cells.
Nat.Chem.Biol., 17, 2021
|
|
8BUH
 
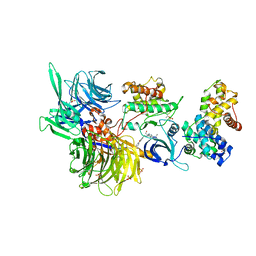 | | Structure of DDB1 bound to WX3-engaged CDK12-cyclin K | | 分子名称: | 6-[[[2-[[(2~{R})-1-oxidanylbutan-2-yl]amino]-9-propan-2-yl-purin-6-yl]amino]methyl]-3-pyridin-2-yl-1~{H}-pyridin-2-one, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.79 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
4A0B
 
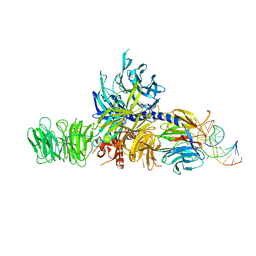 | | Structure of hsDDB1-drDDB2 bound to a 16 bp CPD-duplex (pyrimidine at D-1 position) at 3.8 A resolution (CPD 4) | | 分子名称: | 5'-D(*CP*CP*TP*GP*CP*TP*CP*CP*TP*TP*TP*CP*AP*CP*CP*C)-3', 5'-D(*DGP*GP*TP*GP*AP*AP*AP*(TTD)P*AP*GP*CP*AP*GP*DGP)-3', DNA DAMAGE-BINDING PROTEIN 1, ... | | 著者 | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | 登録日 | 2011-09-08 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
8BUC
 
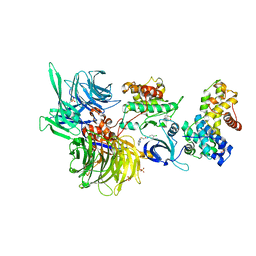 | | Structure of DDB1 bound to dCeMM3-engaged CDK12-cyclin K | | 分子名称: | 2-(1~{H}-benzimidazol-2-ylsulfanyl)-~{N}-(5-chloranylpyridin-2-yl)ethanamide, CITRIC ACID, Cyclin-K, ... | | 著者 | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.85 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
