7N6T
 
 | |
2O4L
 
 | | Crystal Structure of HIV-1 Protease (Q7K, I50V) in Complex with Tipranavir | | 分子名称: | CHLORIDE ION, GLYCEROL, N-(3-{(1R)-1-[(6R)-4-HYDROXY-2-OXO-6-PHENETHYL-6-PROPYL-5,6-DIHYDRO-2H-PYRAN-3-YL]PROPYL}PHENYL)-5-(TRIFLUOROMETHYL)-2-PYRIDINESULFONAMIDE, ... | | 著者 | Armstrong, A.A, Muzammil, S, Jakalian, A, Bonneau, P.R, Schmelmer, V, Freire, E, Amzel, L.M. | | 登録日 | 2006-12-04 | | 公開日 | 2006-12-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations.
J.Virol., 81, 2007
|
|
2QD8
 
 | | HIV-1 Protease Mutant I84V with potent Antiviral inhibitor GRL-98065 | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | 著者 | Wang, Y.F, Tie, Y, Boross, P.I, Tozser, J, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | 登録日 | 2007-06-20 | | 公開日 | 2008-04-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease.
J.Med.Chem., 50, 2007
|
|
8ESX
 
 | | HIV protease in complex with benzoxaborolone analog of darunavir | | 分子名称: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Windsor, I.W, Graham, B.J, Raines, R.T. | | 登録日 | 2022-10-15 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Inhibition of HIV-1 Protease by a Boronic Acid with High Oxidative Stability.
Acs Med.Chem.Lett., 14, 2023
|
|
8ESY
 
 | | D30N mutant HIV protease in complex with benzoxaborolone analog of darunavir | | 分子名称: | CHLORIDE ION, GLYCEROL, Protease, ... | | 著者 | Windsor, I.W, Graham, B.J, Raines, R.T. | | 登録日 | 2022-10-15 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Inhibition of HIV-1 Protease by a Boronic Acid with High Oxidative Stability.
Acs Med.Chem.Lett., 14, 2023
|
|
2BB9
 
 | | Structure of HIV1 protease and AKC4p_133a complex. | | 分子名称: | 2-ETHOXYETHYL (1S,2S)-3-{(2S)-4-[(3AS,8S,8AR)-2-OXO-3,3A,8,8A-TETRAHYDRO-2H-INDENO[1,2-D][1,3]OXAZOL-8-YL]-2-BENZYL-3-OXO-2,3-DIHYDRO-1H-PYRROL-2-YL}-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | 著者 | Smith III, A.B, Charnley, A.K, Kuo, L.C, Munshi, S. | | 登録日 | 2005-10-17 | | 公開日 | 2005-11-15 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Design, synthesis, and biological evaluation of monopyrrolinone-based HIV-1 protease inhibitors possessing augmented P2' side chains
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3TKG
 
 | | crystal structure of HIV model protease precursor/saquinavir complex | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, GLYCEROL, ... | | 著者 | Agniswamy, J, Sayer, J, Weber, I, Louis, J. | | 登録日 | 2011-08-26 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Terminal interface conformations modulate dimer stability prior to amino terminal autoprocessing of HIV-1 protease.
Biochemistry, 51, 2012
|
|
6DJ2
 
 | | HIV-1 protease with single mutation L76V in complex with Lopinavir | | 分子名称: | CHLORIDE ION, HIV-1 protease, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, ... | | 著者 | Wang, Y.-F, Wong-Sam, A.E, Zhang, Y, Weber, I.T. | | 登録日 | 2018-05-24 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease.
ACS Omega, 3, 2018
|
|
4U1J
 
 | | HLA class I micropolymorphisms determine peptide-HLA landscape and dictate differential HIV-1 escape through identical epitopes | | 分子名称: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GAG protein, ... | | 著者 | Rizkallah, P.J, Cole, D.K, Fuller, A, Sewell, A.K. | | 登録日 | 2014-07-15 | | 公開日 | 2015-04-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | A molecular switch in immunodominant HIV-1-specific CD8 T-cell epitopes shapes differential HLA-restricted escape.
Retrovirology, 12, 2015
|
|
4U7V
 
 | | Structure of wild-type HIV protease in complex with degraded photosensitive inhibitor | | 分子名称: | BETA-MERCAPTOETHANOL, N-[(2S,4S,5S)-4-hydroxy-1,6-diphenyl-5-{[(1,3-thiazol-5-ylmethoxy)carbonyl]amino}hexan-2-yl]-L-valinamide, V-1 protease | | 著者 | Pachl, P, Rezacova, P, Schimer, J. | | 登録日 | 2014-07-31 | | 公開日 | 2015-03-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Triggering HIV polyprotein processing by light using rapid photodegradation of a tight-binding protease inhibitor.
Nat Commun, 6, 2015
|
|
3UCB
 
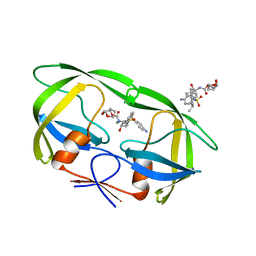 | | Crystal Structure of Multidrug Resistant HIV-1 Protease Clinical Isolate PR20 in Complex with Darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | 著者 | Agniswamy, J, Chen-Hsiang, S, Aniana, A, Sayer, J.M, Louis, J.M, Weber, I.T. | | 登録日 | 2011-10-26 | | 公開日 | 2012-03-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | HIV-1 protease with 20 mutations exhibits extreme resistance to clinical inhibitors through coordinated structural rearrangements.
Biochemistry, 51, 2012
|
|
3QN8
 
 | | HIV-1 protease (mutant Q7K L33I L63I) in complex with a novel inhibitor | | 分子名称: | (4aS,7aS)-1,4-bis(3-hydroxybenzyl)hexahydro-1H-pyrrolo[3,4-b]pyrazine-2,3-dione, CHLORIDE ION, Protease | | 著者 | Lindemann, I, Heine, A, Klebe, G. | | 登録日 | 2011-02-08 | | 公開日 | 2012-02-08 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.382 Å) | | 主引用文献 | Novel inhibitors for HIV-1 protease
To be Published
|
|
7N6V
 
 | |
3QIH
 
 | | HIV-1 protease (mutant Q7K L33I L63I) in complex with a novel inhibitor | | 分子名称: | (4aS,7aS)-1,4-bis(3-hydroxybenzyl)hexahydro-1H-pyrrolo[3,4-b]pyrazine-2,3-dione, CHLORIDE ION, Protease, ... | | 著者 | Lindemann, I, Heine, A, Klebe, G. | | 登録日 | 2011-01-27 | | 公開日 | 2012-02-01 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Novel inhibitors for HIV-1 protease
To be Published
|
|
4FM6
 
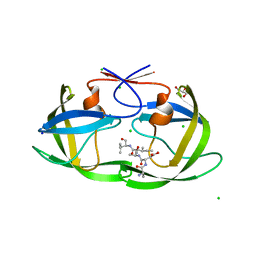 | |
1SDV
 
 | | Crystal structures of HIV protease V82A and L90M mutants reveal changes in indinavir binding site. | | 分子名称: | CHLORIDE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, protease RETROPEPSIN | | 著者 | Mahalingam, B, Wang, Y.-F, Boross, P.I, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | 登録日 | 2004-02-14 | | 公開日 | 2004-05-25 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structures of HIV protease V82A and L90M
mutants reveal changes in the indinavir-binding site
Eur.J.Biochem., 271, 2004
|
|
3QIO
 
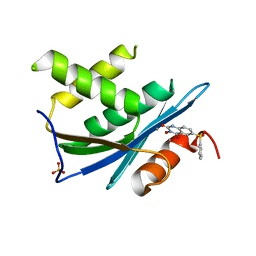 | | Crystal Structure of HIV-1 RNase H with engineered E. coli loop and N-hydroxy quinazolinedione inhibitor | | 分子名称: | 3-hydroxy-6-(phenylsulfonyl)quinazoline-2,4(1H,3H)-dione, Gag-Pol polyprotein,Ribonuclease HI,Gag-Pol polyprotein, MANGANESE (II) ION, ... | | 著者 | Lansdon, E.B, Liu, Q. | | 登録日 | 2011-01-27 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.4011 Å) | | 主引用文献 | Structural and Binding Analysis of Pyrimidinol Carboxylic Acid and N-Hydroxy Quinazolinedione HIV-1 RNase H Inhibitors.
Antimicrob.Agents Chemother., 55, 2011
|
|
2QNP
 
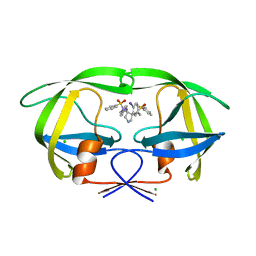 | | HIV-1 Protease in complex with a iodo decorated pyrrolidine-based inhibitor | | 分子名称: | CHLORIDE ION, Gag-Pol polyprotein (Pr160Gag-Pol), N,N'-(3S,4S)-pyrrolidine-3,4-diylbis[N-(4-iodobenzyl)benzenesulfonamide] | | 著者 | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | 登録日 | 2007-07-19 | | 公開日 | 2008-04-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Structure-Guided Design of C2-Symmetric HIV-1 Protease Inhibitors Based on a Pyrrolidine Scaffold.
J.Med.Chem., 51, 2008
|
|
3TOF
 
 | | HIV-1 Protease - Epoxydic Inhibitor Complex (pH 6 - Orthorombic Crystal form P212121) | | 分子名称: | (S)-N-((1R,2S)-1-((2R,3R)-3-benzyloxiran-2-yl)-1-hydroxy-3-phenylpropan-2-yl)-3-methyl-2-(2-phenoxyacetamido)butanamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Geremia, S, Olajuyigbe, F.M, Ajele, J.O, Demitri, N, Randaccio, L, Wuerges, J, Benedetti, L, Campaner, P, Berti, F. | | 登録日 | 2011-09-05 | | 公開日 | 2012-08-15 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Developing HIV-1 Protease Inhibitors through Stereospecific Reactions in Protein Crystals.
Molecules, 21, 2016
|
|
2PK6
 
 | | Crystal Structure of HIV-1 Protease (Q7K, L33I, L63I) in Complex with KNI-10033 | | 分子名称: | (4R)-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-3-[(2S,3S)-2-hydroxy-3-({N-[(isoquinolin-5-yloxy)acetyl]-S-methyl- L-cysteinyl}amino)-4-phenylbutanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Protease | | 著者 | Armstrong, A.A, Lafont, V, Kiso, Y, Freire, E, Amzel, L.M. | | 登録日 | 2007-04-17 | | 公開日 | 2007-05-08 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Compensating enthalpic and entropic changes hinder binding affinity optimization.
Chem.Biol.Drug Des., 69, 2007
|
|
3QBF
 
 | |
3UFN
 
 | | Crystal Structure of Multidrug Resistant HIV-1 Protease Clinical Isolate PR20 in Complex with Saquinavir | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, HIV-1 protease | | 著者 | Agniswamy, J, Chen-Hsiang, S, Aniana, A, Sayer, J.M, Louis, J.M, Weber, I.T. | | 登録日 | 2011-11-01 | | 公開日 | 2012-03-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | HIV-1 protease with 20 mutations exhibits extreme resistance to clinical inhibitors through coordinated structural rearrangements.
Biochemistry, 51, 2012
|
|
3PWR
 
 | | HIV-1 Protease Mutant L76V complexed with Saquinavir | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, GLYCEROL, ... | | 著者 | Zhang, Y, Weber, I.T. | | 登録日 | 2010-12-08 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The L76V Drug Resistance Mutation Decreases the Dimer Stability and Rate of Autoprocessing of HIV-1 Protease by Reducing Internal Hydrophobic Contacts.
Biochemistry, 50, 2011
|
|
3QP0
 
 | | HIV-1 protease (mutant Q7K L33I L63I) in complex with a novel inhibitor | | 分子名称: | (4aS,7aS)-1,4-bis[3-(hydroxymethyl)benzyl]hexahydro-1H-pyrrolo[3,4-b]pyrazine-2,3-dione, CHLORIDE ION, Protease | | 著者 | Lindemann, I, Heine, A, Klebe, G. | | 登録日 | 2011-02-11 | | 公開日 | 2012-02-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Novel inhibitors for HIV-1 protease
To be Published
|
|
3ST5
 
 | | Crystal structure of wild-type HIV-1 protease with C3-Substituted Hexahydrocyclopentafuranyl Urethane as P2-Ligand, GRL-0489A | | 分子名称: | (3R,3aR,5R,6aR)-3-hydroxyhexahydro-2H-cyclopenta[b]furan-5-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Protease | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2011-07-08 | | 公開日 | 2011-08-17 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Design of HIV-1 Protease Inhibitors with C3-Substituted Hexahydrocyclopentafuranyl Urethanes as P2-Ligands: Synthesis, Biological Evaluation, and Protein-Ligand X-ray Crystal Structure.
J.Med.Chem., 54, 2011
|
|
