5HVG
 
 | | Crystal Structure of Thrombin-activatable Fibrinolysis Inhibitor in Complex with an Inhibitory Nanobody (VHH-a204) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Zhou, X, Weeks, S.D, Strelkov, S.V, Declerck, P.J. | | Deposit date: | 2016-01-28 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Elucidation of the molecular mechanisms of two nanobodies that inhibit thrombin-activatable fibrinolysis inhibitor activation and activated thrombin-activatable fibrinolysis inhibitor activity.
J.Thromb.Haemost., 14, 2016
|
|
3CJZ
 
 | | Effects of N2,N2-dimethylguanosine on RNA structure and stability: crystal structure of an RNA duplex with tandem m22G:A pairs | | Descriptor: | RNA (5'-R(*GP*GP*AP*CP*GP*(M2G)P*AP*CP*GP*UP*CP*CP*U)-3') | | Authors: | Pallan, P.S, Kreutz, C, Bosio, S, Micura, R, Egli, M. | | Deposit date: | 2008-03-14 | | Release date: | 2008-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effects of N2,N2-dimethylguanosine on RNA structure and stability: crystal structure of an RNA duplex with tandem m2 2G:A pairs.
Rna, 14, 2008
|
|
5I32
 
 | | Ammonia permeable aquaporin AtTIP2;1 | | Descriptor: | Aquaporin TIP2-1 | | Authors: | Kirscht, A, Nissen, P, Kjellbom, P, Gourdon, P, Johanson, U. | | Deposit date: | 2016-02-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal Structure of an Ammonia-Permeable Aquaporin.
Plos Biol., 14, 2016
|
|
5A8G
 
 | | Crystal structure of the wild-type Staphylococcus aureus N- acetylneurminic acid lyase in complex with fluoropyruvate | | Descriptor: | N-ACETYLNEURAMINATE LYASE | | Authors: | Stockwell, J, Daniels, A.D, Windle, C.L, Harman, T, Woodhall, T, Trinh, C.H, Lebel, T, Pearson, A.R, Mulholland, K, Berry, A, Nelson, A. | | Deposit date: | 2015-07-15 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Evaluation of Fluoropyruvate as Nucleophile in Reactions Catalysed by N-Acetyl Neuraminic Acid Lyase Variants: Scope, Limitations and Stereoselectivity.
Org.Biomol.Chem., 14, 2016
|
|
5KLY
 
 | |
5KM0
 
 | |
5KZS
 
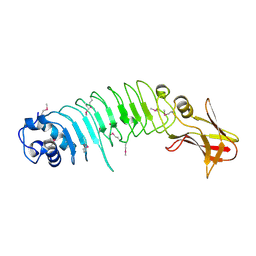 | | Listeria monocytogenes internalin-like protein lmo2027 | | Descriptor: | Putative cell surface protein, similar to internalin proteins | | Authors: | Light, S.H, Nocadello, S, Minasov, G, Cardona-Correa, A, Kwon, K, Faralla, C, Bakardjiev, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-25 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Listeria monocytogenes InlP interacts with afadin and facilitates basement membrane crossing.
Plos Pathog., 14, 2018
|
|
3DD2
 
 | | Crystal structure of an RNA aptamer bound to human thrombin | | Descriptor: | ACETIC ACID, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Long, S.B, Sullenger, B.A. | | Deposit date: | 2008-06-04 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an RNA aptamer bound to thrombin.
Rna, 14, 2008
|
|
5KUD
 
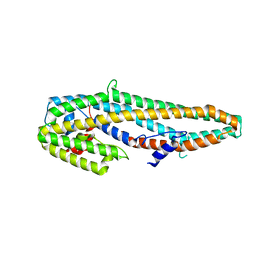 | | Crystal structure of full length Cry6Aa | | Descriptor: | Pesticidal crystal protein Cry6Aa | | Authors: | Kelker, M.S, Xu, X, Lee, M, Chan, M, Hung, S, Dementiev, K, Chikwana, V.M, Hey, T, Narva, K. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The pesticidal Cry6Aa toxin from Bacillus thuringiensis is structurally similar to HlyE-family alpha pore-forming toxins.
Bmc Biol., 14, 2016
|
|
5L16
 
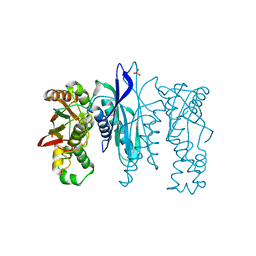 | | Crystal Structure of N-terminus truncated selenophosphate synthetase from Leishmania major | | Descriptor: | Putative selenophosphate synthetase, SULFATE ION | | Authors: | Faim, L.M, Silva, I.R, Pereira, H.M, Dias, M.B, Silva, M.T.A, Brandao-Neto, J, Thiemann, O.H. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Trypanosomatid selenophosphate synthetase structure, function and interaction with selenocysteine lyase.
Plos Negl Trop Dis, 14, 2020
|
|
6ACK
 
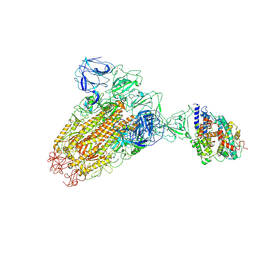 | |
5AXE
 
 | | Crystal Structure Analysis of DNA Duplexes containing sulfoamide-bridged nucleic acid (SuNA-NH) | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(LSH)P*AP*CP*GP*C)-3') | | Authors: | Mitsuoka, Y, Aoyama, H, Kugimiya, A, Fujimura, Y, Yamamoto, T, Waki, R, Wada, F, Tahara, S, Sawamura, M, Noda, M, Hari, Y, Obika, S. | | Deposit date: | 2015-07-28 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Effect of an N-substituent in sulfonamide-bridged nucleic acid (SuNA) on hybridization ability and duplex structure.
Org.Biomol.Chem., 14, 2016
|
|
6AAE
 
 | | Crystal structure of Chloramphenicol-Metabolizaing Enzyme EstDL136 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Esterase, PENTAETHYLENE GLYCOL | | Authors: | Kim, S.H, Kang, P.A, Han, K.T, Lee, S.W, Rhee, S.K. | | Deposit date: | 2018-07-18 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Crystal structure of chloramphenicol-metabolizing enzyme EstDL136 from a metagenome.
PLoS ONE, 14, 2019
|
|
6ABQ
 
 | | Crystal structure of transcription factor from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, PadR family transcriptional regulator | | Authors: | Lee, C, Hong, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based molecular characterization and regulatory mechanism of the LftR transcription factor from Listeria monocytogenes: Conformational flexibilities and a ligand-induced regulatory mechanism.
Plos One, 14, 2019
|
|
1MVP
 
 | |
1N27
 
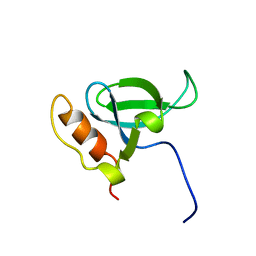 | | Solution structure of the PWWP domain of mouse Hepatoma-derived growth factor, related protein 3 | | Descriptor: | Hepatoma-derived growth factor, related protein 3 | | Authors: | Nameki, N, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-10-22 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PWWP domain of the hepatoma-derived growth factor family.
Protein Sci., 14, 2005
|
|
5SBG
 
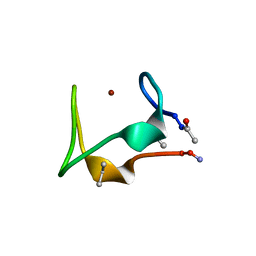 | |
1ST0
 
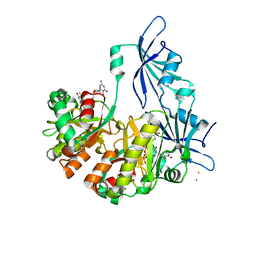 | | Structure of DcpS bound to m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, YTTRIUM (III) ION, mRNA decapping enzyme | | Authors: | Gu, M, Fabrega, C, Liu, S.W, Liu, H, Kiledjian, M, Lima, C.D. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the structure, mechanism, and regulation of scavenger mRNA decapping activity
Mol.Cell, 14, 2004
|
|
1NJR
 
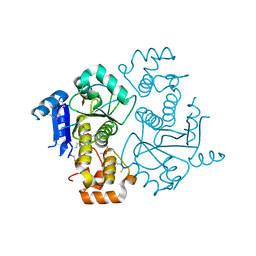 | | Crystal structure of yeast ymx7, an ADP-ribose-1''-monophosphatase | | Descriptor: | 32.1 kDa protein in ADH3-RCA1 intergenic region, Xylitol | | Authors: | Kumaran, D, Eswaramoorthy, S, Studier, F.W, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-01-02 | | Release date: | 2004-08-17 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of ADP-ribose-1''-monophosphatase (Appr-1''-pase), a ubiquitous cellular processing enzyme
Protein Sci., 14, 2005
|
|
1T29
 
 | | Crystal structure of the BRCA1 BRCT repeats bound to a phosphorylated BACH1 peptide | | Descriptor: | BACH1 phosphorylated peptide, Breast cancer type 1 susceptibility protein | | Authors: | Shiozaki, E.N, Gu, L, Yan, N, Shi, Y. | | Deposit date: | 2004-04-20 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the BRCT repeats of BRCA1 bound to a BACH1 phosphopeptide: implications for signaling.
Mol.Cell, 14, 2004
|
|
1T7K
 
 | | Crystal Structure of HIV Protease complexed with Arylsulfonamide azacyclic urea | | Descriptor: | 3-({5-BENZYL-6-HYDROXY-2,4-BIS-(4-HYDROXY-BENZYL)-3-OXO-[1,2,4]-TRIAZEPANE-1-SULFONYL)-BENZONITRILE, Pol polyprotein [Contains: Protease (Retropepsin)] | | Authors: | Huang, P.P, Randolph, J.T, Klein, L.L, Vasavanonda, S, Dekhtyar, T, Stoll, V.S, Kempf, D.J. | | Deposit date: | 2004-05-10 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis and Antiviral Activity of P1' Arylsulfonamide Azacyclic Urea HIV Protease Inhibitors
Bioorg.Med.Chem.Lett., 14, 2004
|
|
6AL7
 
 | | Crystal structure HpiC1 F138S | | Descriptor: | 12-epi-hapalindole C/U synthase, CALCIUM ION | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.687 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
6A5I
 
 | | Pseudocerastes Persicus Trypsin Inhibitor | | Descriptor: | Trypsin Inhibitor | | Authors: | Amininasab, M. | | Deposit date: | 2018-06-23 | | Release date: | 2019-05-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of PPTI, a kunitz-type protein from the venom of Pseudocerastes persicus.
PLoS ONE, 14, 2019
|
|
6ABX
 
 | |
1R5H
 
 | | Crystal Structure of MetAP2 complexed with A320282 | | Descriptor: | MANGANESE (II) ION, Methionine aminopeptidase 2, N'-(2S,3R)-3-AMINO-4-CYCLOHEXYL-2-HYDROXY-BUTANO-N-(4-METHYLPHENYL)HYDRAZIDE | | Authors: | Sheppard, G.S, Wang, J, Kawai, M, BaMaung, N.Y, Craig, R.A, Erickson, S.A, Lynch, L, Patel, J, Yang, F, Searle, X.B, Lou, P, Park, C, Kim, K.H, Henkin, J, Lesniewski, R. | | Deposit date: | 2003-10-10 | | Release date: | 2004-10-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3-Amino-2-hydroxyamides and related compounds as inhibitors of methionine aminopeptidase-2.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
