5IE2
 
 | | Crystal structure of a plant enzyme | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fan, M.R, Li, M, Chang, W.R. | | Deposit date: | 2016-02-24 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Arabidopsis thaliana Oxalyl-CoA Synthetase Essential for Oxalate Degradation
Mol Plant, 9, 2016
|
|
7OJX
 
 | | E2 UBE2K covalently linked to donor Ub, acceptor di-Ub, and RING E3 primed for K48-linked Ub chain synthesis | | Descriptor: | 1,1'-ethane-1,2-diylbis(1H-pyrrole-2,5-dione), E3 ubiquitin-protein ligase RNF38, Polyubiquitin-B, ... | | Authors: | Majorek, K.A, Nakasone, M.A, Huang, D.T. | | Deposit date: | 2021-05-17 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of UBE2K-Ub/E3/polyUb reveals mechanisms of K48-linked Ub chain extension.
Nat.Chem.Biol., 18, 2022
|
|
7UXV
 
 | | human triosephosphate isomerase mutant G122R | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Romero, J.M. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of the TPI-Manchester, a thermolabile variant of human triosephosphate isomerase.
Arch.Biochem.Biophys., 761, 2024
|
|
5LCW
 
 | | Cryo-EM structure of the Anaphase-promoting complex/Cyclosome, in complex with the Mitotic checkpoint complex (APC/C-MCC) at 4.2 angstrom resolution | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Alfieri, C, Chang, L, Zhang, Z, Yang, J, Maslen, S, Skehel, M, Barford, D. | | Deposit date: | 2016-06-22 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Molecular basis of APC/C regulation by the spindle assembly checkpoint.
Nature, 536, 2016
|
|
6FZF
 
 | | PPAR mutant complex | | Descriptor: | (2~{S})-3-[4-[2-[methyl(pyridin-2-yl)amino]ethoxy]phenyl]-2-[[2-(phenylcarbonyl)phenyl]amino]propanoic acid, Peroxisome proliferator-activated receptor gamma, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | Authors: | Rochel, N. | | Deposit date: | 2018-03-14 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Recurrent activating mutations of PPAR gamma associated with luminal bladder tumors.
Nat Commun, 10, 2019
|
|
5IE0
 
 | | Crystal structure of a plant enzyme | | Descriptor: | Oxalate--CoA ligase, S,R MESO-TARTARIC ACID | | Authors: | Ran, M.R, Li, M, Chang, W.R. | | Deposit date: | 2016-02-24 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Arabidopsis thaliana Oxalyl-CoA Synthetase Essential for Oxalate Degradation
Mol Plant, 9, 2016
|
|
8TB0
 
 | | Cryo-EM Structure of GPR61-G protein complex stabilized by scFv16 | | Descriptor: | GPR61 fused to dominant negative G alpha S/I N18 chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Lees, J.A, Dias, J.M, Han, S. | | Deposit date: | 2023-06-28 | | Release date: | 2023-10-04 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | An inverse agonist of orphan receptor GPR61 acts by a G protein-competitive allosteric mechanism.
Nat Commun, 14, 2023
|
|
8E97
 
 | | PYD-106-bound Human GluN1a-GluN2C NMDA receptor in splayed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
8E94
 
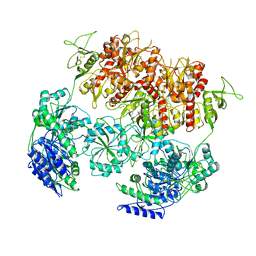 | | PYD-106-bound Human GluN1a-GluN2C NMDA receptor in intact conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
8E93
 
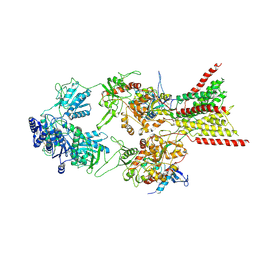 | | D-cycloserine and glutamate bound Human GluN1a-GluN2C NMDA receptor in splayed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
8E92
 
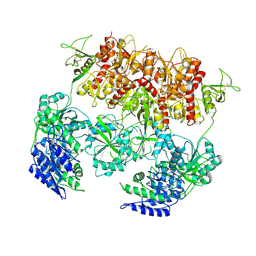 | | D-cycloserine and glutamate bound Human GluN1a-GluN2C NMDA receptor in intact conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
3MNA
 
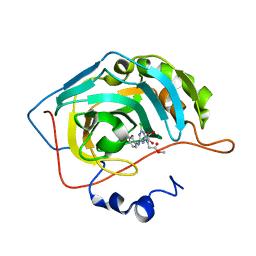 | | The crystal structure of human carbonic anhydrase Ii in complex with a 1,3,5-triazine-substituted benzenesulfonamide inhibitor | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, ZINC ION, ... | | Authors: | Avvaru, B.S, Wagner, J, Robbins, A.H, Mckenna, R. | | Deposit date: | 2010-04-21 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Sulfonamides incorporating 1,3,5-triazine moieties selectively and potently inhibit carbonic anhydrase transmembrane isoforms IX, XII and XIV over cytosolic isoforms I and II: Solution and X-ray crystallographic studies.
Bioorg.Med.Chem., 19, 2011
|
|
6RQI
 
 | | Human Carbonic Anhydrase II in complex with fluorinated benzenesulfonamide | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 3-fluorobenzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The Influence of Varying Fluorination Patterns on the Thermodynamics and Kinetics of Benzenesulfonamide Binding to Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
1NC3
 
 | | Crystal structure of E. coli MTA/AdoHcy nucleosidase complexed with formycin A (FMA) | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2002-12-04 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Escherichia coli 5'-methylthioadenosine/ S-adenosylhomocysteine nucleosidase inhibitor complexes provide insight into the conformational changes required for substrate binding and catalysis.
J.Biol.Chem., 278, 2003
|
|
6GTL
 
 | | Achromobacter cycloclastes copper nitrite reductase at pH 6.0 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Halsted, T.P, Eady, R.R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2018-06-18 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Catalytically important damage-free structures of a copper nitrite reductase obtained by femtosecond X-ray laser and room-temperature neutron crystallography.
Iucrj, 6, 2019
|
|
3HLE
 
 | | Simvastatin Synthase (LovD), from Aspergillus terreus, S5 mutant, S76A mutant, complex with monacolin J acid | | Descriptor: | (3R,5R)-3,5-dihydroxy-7-[(1S,2S,6R,8S,8aR)-8-hydroxy-2,6-dimethyl-1,2,6,7,8,8a-hexahydronaphthalen-1-yl]heptanoic acid, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Transesterase | | Authors: | Sawaya, M.R, Yeates, T.O, Pashkov, I, Gao, X, Tang, Y. | | Deposit date: | 2009-05-27 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Directed evolution and structural characterization of a simvastatin synthase
Chem.Biol., 16, 2009
|
|
4WUQ
 
 | | Crystal structure of human carbonic anhydrase isozyme I with 2,3,5,6-Tetrafluoro-4-piperidin-1-ylbenzenesulfonamide | | Descriptor: | 2,3,5,6-tetrafluoro-4-(piperidin-1-yl)benzenesulfonamide, Carbonic anhydrase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Manakova, E, Smirnov, A, Grazulis, S. | | Deposit date: | 2014-11-03 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Intrinsic thermodynamics of 4-substituted-2,3,5,6-tetrafluorobenzenesulfonamide binding to carbonic anhydrases by isothermal titration calorimetry.
Biophys.Chem., 205, 2015
|
|
6DLZ
 
 | | Open state GluA2 in complex with STZ after micelle signal subtraction | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Vassilevski, A.A, Sobolevsky, A.I. | | Deposit date: | 2018-06-04 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors.
Neuron, 99, 2018
|
|
2GGK
 
 | |
6H0F
 
 | | Structure of DDB1-CRBN-pomalidomide complex bound to IKZF1(ZF2) | | Descriptor: | DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1, DNA-binding protein Ikaros, Protein cereblon, ... | | Authors: | Petzold, G, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2018-07-09 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN.
Science, 362, 2018
|
|
9C8L
 
 | |
9C8P
 
 | |
6ZQH
 
 | | Yeast Uba1 in complex with ubiquitin | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | Development of ADPribosyl Ubiquitin Analogues to Study Enzymes Involved in Legionella Infection.
Chemistry, 27, 2021
|
|
6H0G
 
 | | Structure of the DDB1-CRBN-pomalidomide complex bound to ZNF692(ZF4) | | Descriptor: | DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1,DDB1 (DNA damage binding protein 1),DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | Authors: | Bunker, R.D, Petzold, G, Thoma, N.H. | | Deposit date: | 2018-07-09 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.25 Å) | | Cite: | Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN.
Science, 362, 2018
|
|
8DN2
 
 | |
