8J1E
 
 | | AtSLAC1 in open state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2023-04-12 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
8J05
 
 | |
8J03
 
 | |
8J04
 
 | | Human KCNQ2-CaM-HN37 complex in the presence of PIP2 | | Descriptor: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, methyl N-[4-[(4-fluorophenyl)methyl-prop-2-ynyl-amino]-2,6-dimethyl-phenyl]carbamate | | Authors: | Ma, D, Li, X, Guo, J. | | Deposit date: | 2023-04-09 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
1QPI
 
 | | CRYSTAL STRUCTURE OF TETRACYCLINE REPRESSOR/OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*CP*CP*TP*AP*TP*CP*AP*AP*TP*GP*AP*TP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*AP*TP*CP*AP*TP*TP*GP*AP*TP*AP*GP*G)-3'), IMIDAZOLE, ... | | Authors: | Orth, P, Schnappinger, D, Hillen, W, Saenger, W, Hinrichs, W. | | Deposit date: | 1999-05-25 | | Release date: | 2000-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system.
Nat.Struct.Biol., 7, 2000
|
|
1H0I
 
 | | Complex of a chitinase with the natural product cyclopentapeptide argifin from Gliocladium | | Descriptor: | ARGIFIN, CHITINASE B, GLYCEROL, ... | | Authors: | Houston, D.R, Shiomi, K, Arai, N, Omura, S, Peter, M.G, Turberg, A, Synstad, B, Eijsink, V.G.H, Aalten, D.M.F. | | Deposit date: | 2002-06-19 | | Release date: | 2002-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution Inhibited Complexes of a Chitinase with Natural Product Cyclopentapeptides - Peptide Mimicry of a Carbohydrate Substrate
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1QW5
 
 | | Murine inducible nitric oxide synthase oxygenase domain in complex with W1400 inhibitor. | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, N-(3-(AMINOMETHYL)BENZYL)ACETAMIDINE, Nitric oxide synthase, ... | | Authors: | Fedorov, R, Hartmann, E, Ghosh, D.K, Schlichting, I. | | Deposit date: | 2003-08-31 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the specificity of the nitric-oxide synthase inhibitors W1400 and Nomega-propyl-L-Arg for the inducible and neuronal isoforms.
J.Biol.Chem., 278, 2003
|
|
1H9N
 
 | | H119N CARBONIC ANHYDRASE II | | Descriptor: | CARBONIC ANHYDRASE II, ZINC ION | | Authors: | Lesburg, C.A, Christianson, D.W. | | Deposit date: | 1997-05-29 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Histidine --> carboxamide ligand substitutions in the zinc binding site of carbonic anhydrase II alter metal coordination geometry but retain catalytic activity.
Biochemistry, 36, 1997
|
|
1QWO
 
 | | Crystal structure of a phosphorylated phytase from Aspergillus fumigatus, revealing the structural basis for its heat resilience and catalytic pathway | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, phytase | | Authors: | Xiang, T, Liu, Q, Deacon, A.M, Koshy, M, Kriksunov, I.A, Lei, X.G, Hao, Q, Thiel, D.J. | | Deposit date: | 2003-09-03 | | Release date: | 2004-06-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Heat-resilient Phytase from Aspergillus fumigatus, Carrying a Phosphorylated Histidine
J.Mol.Biol., 339, 2004
|
|
1H9Q
 
 | | H119Q CARBONIC ANHYDRASE II | | Descriptor: | CARBONIC ANHYDRASE II, ZINC ION | | Authors: | Lesburg, C.A, Christianson, D.W. | | Deposit date: | 1997-05-29 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Histidine --> carboxamide ligand substitutions in the zinc binding site of carbonic anhydrase II alter metal coordination geometry but retain catalytic activity.
Biochemistry, 36, 1997
|
|
8J02
 
 | | Human KCNQ2(F104A)-CaM-PIP2-CBD complex in state II | | Descriptor: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate, ... | | Authors: | Ma, D, Li, X, Guo, J. | | Deposit date: | 2023-04-09 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
8J01
 
 | | Human KCNQ2-CaM in complex with CBD and PIP2 | | Descriptor: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate, ... | | Authors: | Ma, D, Li, X, Guo, J. | | Deposit date: | 2023-04-09 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
8J46
 
 | | Human Consensus Olfactory Receptor OR52c in apo state, OR52c-bRIL | | Descriptor: | Olfactory receptor OR52c,Soluble cytochrome b562 | | Authors: | Choi, C.W, Bae, J, Choi, H.-J, Kim, J. | | Deposit date: | 2023-04-19 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Understanding the molecular mechanisms of odorant binding and activation of the human OR52 family.
Nat Commun, 14, 2023
|
|
1QV0
 
 | | Atomic resolution structure of obelin from Obelia longissima | | Descriptor: | C2-HYDROPEROXY-COELENTERAZINE, COBALT (II) ION, GLYCEROL, ... | | Authors: | Liu, Z.J, Vysotski, E.S, Deng, L, Lee, J, Rose, J, Wang, B.C. | | Deposit date: | 2003-08-26 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution structure of obelin: soaking with calcium enhances electron density of the second oxygen atom substituted at the C2-position of coelenterazine.
Biochem.Biophys.Res.Commun., 311, 2003
|
|
1HGB
 
 | | HIGH RESOLUTION CRYSTAL STRUCTURES AND COMPARISONS OF T STATE DEOXYHAEMOGLOBIN AND TWO LIGANDED T-STATE HAEMOGLOBINS: T(ALPHA-OXY)HAEMOGLOBIN AND T(MET)HAEMOGLOBIN | | Descriptor: | HEMOGLOBIN (AQUO MET) (ALPHA CHAIN), HEMOGLOBIN (AQUO MET) (BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Liddington, R, Derewenda, Z, Dodson, E, Hubbard, R, Dodson, G. | | Deposit date: | 1991-10-31 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High resolution crystal structures and comparisons of T-state deoxyhaemoglobin and two liganded T-state haemoglobins: T(alpha-oxy)haemoglobin and T(met)haemoglobin.
J.Mol.Biol., 228, 1992
|
|
1HGA
 
 | | HIGH RESOLUTION CRYSTAL STRUCTURES AND COMPARISONS OF T STATE DEOXYHAEMOGLOBIN AND TWO LIGANDED T-STATE HAEMOGLOBINS: T(ALPHA-OXY)HAEMOGLOBIN AND T(MET)HAEMOGLOBIN | | Descriptor: | HEMOGLOBIN (DEOXY) (ALPHA CHAIN), HEMOGLOBIN (DEOXY) (BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Liddington, R, Derewenda, Z, Dodson, E, Hubbard, R, Dodson, G. | | Deposit date: | 1991-10-31 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High resolution crystal structures and comparisons of T-state deoxyhaemoglobin and two liganded T-state haemoglobins: T(alpha-oxy)haemoglobin and T(met)haemoglobin.
J.Mol.Biol., 228, 1992
|
|
8JCU
 
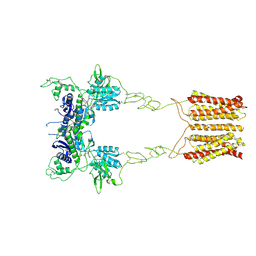 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495 (dimerization mode I) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
1HCO
 
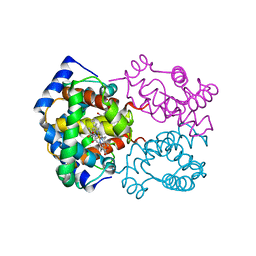 | |
1HHO
 
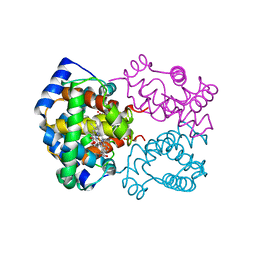 | | STRUCTURE OF HUMAN OXYHAEMOGLOBIN AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | HEMOGLOBIN A (OXY) (ALPHA CHAIN), HEMOGLOBIN A (OXY) (BETA CHAIN), OXYGEN MOLECULE, ... | | Authors: | Shaanan, B. | | Deposit date: | 1983-06-10 | | Release date: | 1983-10-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of human oxyhaemoglobin at 2.1 A resolution.
J.Mol.Biol., 171, 1983
|
|
8JCX
 
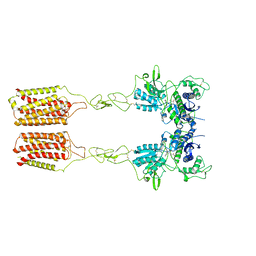 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495 and NAM563 (dimerization mode II) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
1HGC
 
 | | HIGH RESOLUTION CRYSTAL STRUCTURES AND COMPARISONS OF T STATE DEOXYHAEMOGLOBIN AND TWO LIGANDED T-STATE HAEMOGLOBINS: T(ALPHA-OXY)HAEMOGLOBIN AND T(MET)HAEMOGLOBIN | | Descriptor: | HEMOGLOBIN (DEOXY) (BETA CHAIN), HEMOGLOBIN (OXY) (ALPHA CHAIN), OXYGEN MOLECULE, ... | | Authors: | Liddington, R, Derewenda, Z, Dodson, E, Hubbard, R, Dodson, G. | | Deposit date: | 1991-10-31 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High resolution crystal structures and comparisons of T-state deoxyhaemoglobin and two liganded T-state haemoglobins: T(alpha-oxy)haemoglobin and T(met)haemoglobin.
J.Mol.Biol., 228, 1992
|
|
8JD0
 
 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of NAM563 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(1-methylpyrazol-4-yl)-7-[[(2~{S})-2-(trifluoromethyl)morpholin-4-yl]methyl]quinoline-2-carboxamide, CHOLESTEROL, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
1NQ9
 
 | | Crystal Structure of Antithrombin in the Pentasaccharide-Bound Intermediate State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3,4-di-O-methyl-2,6-di-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-beta-D-glucopyranuronic acid-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-3-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2,3,6-tri-O-sulfo-alpha-D-glucopyranoside, ... | | Authors: | Huntington, J.A, Johnson, D.J.D. | | Deposit date: | 2003-01-21 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Antithrombin in a Heparin-Bound Intermediate State
Biochemistry, 42, 2003
|
|
8JCY
 
 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495, NAM563, and LY2389575 (dimerization mode I) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
1NU1
 
 | | Crystal Structure of Mitochondrial Cytochrome bc1 Complexed with 2-nonyl-4-hydroxyquinoline N-oxide (NQNO) | | Descriptor: | 2-NONYL-4-HYDROXYQUINOLINE N-OXIDE, Cytochrome b, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Gao, X, Wen, X, Esser, L, Quinn, B, Yu, L, Yu, C.-A, Xia, D. | | Deposit date: | 2003-01-30 | | Release date: | 2003-10-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the quinone reduction in the bc(1) complex: a comparative analysis of crystal structures of mitochondrial cytochrome bc(1) with bound substrate and inhibitors at the Q(i) site
Biochemistry, 42, 2003
|
|
