4LE5
 
 | |
2GPS
 
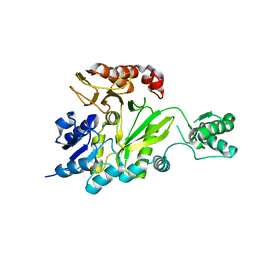 | | Crystal Structure of the Biotin Carboxylase Subunit, E23R mutant, of Acetyl-CoA Carboxylase from Escherichia coli. | | Descriptor: | Biotin carboxylase | | Authors: | Shen, Y, Chou, C.Y, Chang, G.G, Tong, L. | | Deposit date: | 2006-04-18 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Is dimerization required for the catalytic activity of bacterial biotin carboxylase?
Mol.Cell, 22, 2006
|
|
5FO1
 
 | |
5FO0
 
 | |
2EQA
 
 | | Crystal Structure of the hypothetical Sua5 protein from Sulfolobus tokodaii | | Descriptor: | ADENOSINE MONOPHOSPHATE, Hypothetical protein ST1526, MAGNESIUM ION | | Authors: | Agari, Y, Shinkai, A, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2008-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of a hypothetical Sua5 protein from Sulfolobus tokodaii strain 7
Proteins, 70, 2008
|
|
3KFU
 
 | | Crystal structure of the transamidosome | | Descriptor: | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, Glutamyl-tRNA(Gln) amidotransferase subunit A, Glutamyl-tRNA(Gln) amidotransferase subunit C, ... | | Authors: | Blaise, M, Bailly, M, Frechin, M, Thirup, S, Becker, H.D, Kern, D. | | Deposit date: | 2009-10-28 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a transfer-ribonucleoprotein particle that promotes asparagine formation.
Embo J., 29, 2010
|
|
3JRW
 
 | | Phosphorylated BC domain of ACC2 | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Cho, Y.S, Lee, J.I, Shin, D, Kim, H.T, Lee, T.G, Heo, Y.S. | | Deposit date: | 2009-09-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism for the regulation of human ACC2 through phosphorylation by AMPK
Biochem.Biophys.Res.Commun., 391, 2010
|
|
3JRX
 
 | | Crystal structure of the BC domain of ACC2 in complex with soraphen A | | Descriptor: | Acetyl-CoA carboxylase 2, SORAPHEN A | | Authors: | Cho, Y.S, Lee, J.I, Shin, D, Kim, H.T, Lee, T.G, Heo, Y.S. | | Deposit date: | 2009-09-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism for the regulation of human ACC2 through phosphorylation by AMPK.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
6RJS
 
 | | Inter-dimeric interface controls function and stability of S-methionine adenosyltransferase from U. urealiticum | | Descriptor: | Methionine adenosyltransferase | | Authors: | Shahar, A, Zarivach, R, Bershtein, S, Kleiner, D, Shmulevich, F. | | Deposit date: | 2019-04-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The interdimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase.
J.Mol.Biol., 431, 2019
|
|
6RK5
 
 | | Inter-dimeric interface controls function and stability of S-methionine adenosyltransferase from U. urealiticum | | Descriptor: | Methionine adenosyltransferase | | Authors: | Shahar, A, Zarivach, R, Bershtein, S, Kleiner, D, Shmulevich, F. | | Deposit date: | 2019-04-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The interdimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase.
J.Mol.Biol., 431, 2019
|
|
6RK7
 
 | | Inter-dimeric interface controls function and stability of S-methionine adenosyltransferase from U. urealiticum | | Descriptor: | CHLORIDE ION, Methionine adenosyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Shahar, A, Zarivach, R, Bershtein, S, Kleiner, D, Shmulevich, F. | | Deposit date: | 2019-04-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The interdimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase.
J.Mol.Biol., 431, 2019
|
|
7D9K
 
 | |
7D9Y
 
 | |
6RKC
 
 | | Inter-dimeric interface controls function and stability of S-methionine adenosyltransferase from U. urealiticum | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, MAGNESIUM ION, Methionine adenosyltransferase, ... | | Authors: | Shahar, A, Zarivach, R, Bershtein, S, Kleiner, D, Shmulevich, F. | | Deposit date: | 2019-04-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The interdimeric interface controls function and stability of Ureaplasma urealiticum methionine S-adenosyltransferase.
J.Mol.Biol., 431, 2019
|
|
8A8M
 
 | | Structure of the MAPK p38alpha in complex with its activating MAP2K MKK6 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 6, MAGNESIUM ION, Mitogen-activated protein kinase 14, ... | | Authors: | Bowler, M.W, Juyoux, P, Pellegrini, E. | | Deposit date: | 2022-06-23 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Architecture of the MKK6-p38 alpha complex defines the basis of MAPK specificity and activation.
Science, 381, 2023
|
|
8RTY
 
 | | Structure of the F-actin barbed end bound by Cdc12 and profilin (ring complex) at a resolution of 6.3 Angstrom | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Funk, J, Prumbaum, D, Raunser, S, Bieling, P. | | Deposit date: | 2024-01-29 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (6.25 Å) | | Cite: | Molecular mechanism of actin filament elongation by formins.
Science, 384, 2024
|
|
4BUB
 
 | | CRYSTAL STRUCTURE OF MURE LIGASE FROM THERMOTOGA MARITIMA IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, UDP-N-ACETYLMURAMOYL-L-ALANYL-D-GLUTAMATE--LD-LYSINE LIGASE | | Authors: | Favini-Stabile, S, Contreras-Martel, C, Thielens, N, Dessen, A. | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mreb and Murg as Scaffolds for the Cytoplasmic Steps of Peptidoglycan Biosynthesis
Environ.Microbiol., 15, 2013
|
|
4F8E
 
 | | Crystal structure of human PRS1 D52H mutant | | Descriptor: | MAGNESIUM ION, Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | Authors: | Chen, P, Teng, M, Li, X. | | Deposit date: | 2012-05-17 | | Release date: | 2013-05-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | A small disturbance, but a serious disease: the possible mechanism of D52H-mutant of human PRS1 that causes gout
Iubmb Life, 65, 2013
|
|
8COG
 
 | | Human arginylated beta-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Pinto, C.S, Bakker, S.E, Suchenko, A, Hussain, H, Hatano, T, Sampath, K, Chinthalapudi, K, Mishima, M, Balasubramanian, M. | | Deposit date: | 2023-02-28 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.499 Å) | | Cite: | Structure and physiological investigation of human arginylated beta-actin
To Be Published
|
|
8F5D
 
 | | Architecture of the MurE-MurF ligase bacterial cell wall biosynthesis complex | | Descriptor: | Multifunctional fusion protein, SULFATE ION | | Authors: | Shirakawa, K.T, Sala, F.A, Miyachiro, M.M, Job, V, Trindade, D.M, Dessen, A. | | Deposit date: | 2022-11-14 | | Release date: | 2023-06-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Architecture and genomic arrangement of the MurE-MurF bacterial cell wall biosynthesis complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3HMQ
 
 | | 1.9 Angstrom resolution crystal structure of a NAD synthetase (nadE) from Salmonella typhimurium LT2 in complex with NAD(+) | | Descriptor: | NH(3)-dependent NAD(+) synthetase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Halavaty, A.S, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-29 | | Release date: | 2009-06-16 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom resolution crystal structure of a NAD synthetase (nadE) from Salmonella typhimurium LT2 in complex with NAD(+)
To be Published
|
|
3IP4
 
 | | The high resolution structure of GatCAB | | Descriptor: | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit C, Glutamyl-tRNA(Gln) amidotransferase subunit A, ... | | Authors: | Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2009-08-17 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Two distinct regions in Staphylococcus aureus GatCAB guarantee accurate tRNA recognition
Nucleic Acids Res., 38, 2010
|
|
3VNN
 
 | | Crystal Structure of a sub-domain of the nucleotidyltransferase (adenylation) domain of human DNA ligase IV | | Descriptor: | DNA ligase 4 | | Authors: | Ochi, T, Wu, Q, Chirgadze, D.Y, Grossmann, J.G, Bolanos-Garcia, V.M, Blundell, T.L. | | Deposit date: | 2012-01-17 | | Release date: | 2012-06-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Structural insights into the role of domain flexibility in human DNA ligase IV
Structure, 20, 2012
|
|
6YBQ
 
 | | Engineered glycolyl-CoA carboxylase (quintuple mutant) with bound CoA | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, COENZYME A, Propionyl-CoA carboxylase alpha subunit, ... | | Authors: | Schuller, J.M, Schuller, S.K, Zarzycki, J, Scheffen, M, Marchal, D.M, Erb, T.J. | | Deposit date: | 2020-03-17 | | Release date: | 2020-10-28 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (1.96 Å) | | Cite: | A new-to-nature carboxylation module to improve natural and synthetic CO2 fixation
Nat Catal, 2021
|
|
6YBP
 
 | | Propionyl-CoA carboxylase of Methylorubrum extorquens with bound CoA | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, COENZYME A, Propionyl-CoA carboxylase alpha subunit, ... | | Authors: | Schuller, J.M, Schuller, S.K, Zarzycki, J, Scheffen, M, Marchal, D.M, Erb, T.J. | | Deposit date: | 2020-03-17 | | Release date: | 2020-10-28 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | A new-to-nature carboxylation module to improve natural and synthetic CO2 fixation
Nat Catal, 2021
|
|
