6PZ9
 
 | | Cryo-EM structure of the pancreatic beta-cell SUR1 bound to ATP and repaglinide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ATP-sensitive inward rectifier potassium channel 11, ... | | Authors: | Shyng, S.L, Yoshioka, C, Martin, G.M, Sung, M.W. | | Deposit date: | 2019-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Mechanism of pharmacochaperoning in a mammalian K ATP channel revealed by cryo-EM.
Elife, 8, 2019
|
|
1MAY
 
 | | BETA-TRYPSIN PHOSPHONATE INHIBITED | | Descriptor: | BETA-TRYPSIN, CALCIUM ION, [N-(BENZYLOXYCARBONYL)AMINO](4-AMIDINOPHENYL)METHANE-PHOSPHONATE | | Authors: | Bertrand, J, Oleksyszyn, J, Kam, C, Boduszek, B, Presnell, S, Plaskon, R, Suddath, F, Powers, J, Williams, L. | | Deposit date: | 1996-02-06 | | Release date: | 1996-10-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of trypsin and thrombin by amino(4-amidinophenyl)methanephosphonate diphenyl ester derivatives: X-ray structures and molecular models.
Biochemistry, 35, 1996
|
|
6Q1A
 
 | |
5YTC
 
 | | Large fragment of DNA Polymerase I from Thermus aquaticus in a closed ternary complex with the unnatural base M-fC pair with dATP in the active site | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*(92F)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC)P*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Yi, C.Q. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6Q3D
 
 | | Dye type peroxidase Aa from Streptomyces lividans: 235.2 kGy structure | | Descriptor: | Deferrochelatase/peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ebrahim, A, Moreno-Chicano, T, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Appleby, M, Owen, R.L. | | Deposit date: | 2018-12-04 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Dose-resolved serial synchrotron and XFEL structures of radiation-sensitive metalloproteins.
Iucrj, 6, 2019
|
|
6Q5J
 
 | | Crystal structure of a CC-Hex mutant that forms a parallel six-helix coiled coil CC-Hex*-L24E | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CC-Hex*-L24E | | Authors: | Rhys, G.G, Wood, C.W, Beesley, J.L, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5M
 
 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-L24Dab | | Descriptor: | CC-Hex*-L24Dab, SODIUM ION | | Authors: | Wood, C.W, Beesley, J.L, Rhys, G.G, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
5YTF
 
 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base M-fC pair with dA | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(92F)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5YTG
 
 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base I-fC pair with dA | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(94O)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5YTH
 
 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base M-fC pair with dG | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(92F)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*GP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5YJ4
 
 | |
5YAM
 
 | |
5YW9
 
 | |
5YWB
 
 | |
5YWC
 
 | |
5Z2S
 
 | | Crystal structure of DUX4-HD2 domain | | Descriptor: | Double homeobox protein 4 | | Authors: | Dong, X, Zhang, W, Wu, H, Huang, J, Zhang, M, Wang, P, Zhang, H, Chen, Z, Chen, S, Meng, G. | | Deposit date: | 2018-01-03 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of DUX4/IGH-driven transactivation.
Leukemia, 32, 2018
|
|
6DZU
 
 | |
5Z6J
 
 | |
5ZEW
 
 | |
5YW8
 
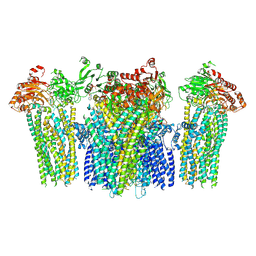 | |
6E5P
 
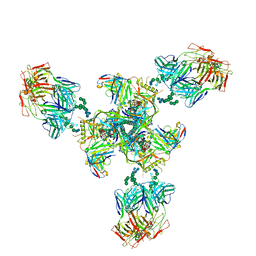 | | Backbone model based on cryo-EM map at 8.5 A of domain-swapped, glycan-reactive, neutralizing antibody 2G12 bound to HIV-1 Env BG505 DS-SOSIP, which was also bound to CD4-binding site antibody VRC03 | | Descriptor: | 2G12 Light chain, 2G12 heavy chain, Envelope glycoprotein gp120, ... | | Authors: | Acharya, P, Kwong, P.D. | | Deposit date: | 2018-07-21 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Structural Survey of Broadly Neutralizing Antibodies Targeting the HIV-1 Env Trimer Delineates Epitope Categories and Characteristics of Recognition.
Structure, 27, 2019
|
|
5Z6Y
 
 | | Structure of sfYFP48S95C66BPA | | Descriptor: | Green fluorescent protein | | Authors: | Wang, J.Y, Wang, J.Y. | | Deposit date: | 2018-01-25 | | Release date: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | structure of sfYFP48S95C66BPA at 1.95 Angstroms resolution
To Be Published
|
|
8QH5
 
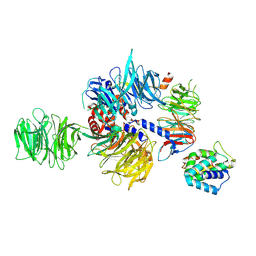 | | CryoEM structure of UVSSA(VHS)-CSA-DDB1-DDA1 | | Descriptor: | DET1- and DDB1-associated protein 1, DNA damage-binding protein 1, DNA excision repair protein ERCC-8, ... | | Authors: | Lee, S.-H, Sixma, T.K. | | Deposit date: | 2023-09-06 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | CryoEM structure of UVSSA(VHS)-CSA-DDB1-DDA1
To Be Published
|
|
8UBR
 
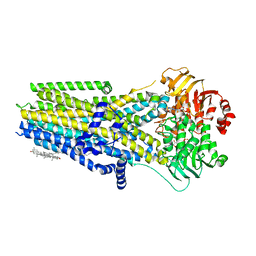 | | Complex of the phosphorylated human cystic fibrosis transmembrane conductance regulator (CFTR) with CFTRinh-172 and ATP/Mg | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 4-[(Z)-{(3M)-4-oxo-2-sulfanylidene-3-[3-(trifluoromethyl)phenyl]-1,3-thiazolidin-5-ylidene}methyl]benzoic acid, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Young, P.G, Fiedorczuk, K, Chen, J. | | Deposit date: | 2023-09-24 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for CFTR inhibition by CFTR inh -172.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5ZC9
 
 | | Crystal structure of the human eIF4A1-ATP analog-RocA-polypurine RNA complex | | Descriptor: | (1R,2R,3S,3aR,8bS)-6,8-dimethoxy-3a-(4-methoxyphenyl)-N,N-dimethyl-1,8b-bis(oxidanyl)-3-phenyl-2,3-dihydro-1H-cyclopenta[b][1]benzofuran-2-carboxamide, Eukaryotic initiation factor 4A-I, MAGNESIUM ION, ... | | Authors: | Iwasaki, W, Takahashi, M, Sakamoto, A, Iwasaki, S, Ito, T. | | Deposit date: | 2018-02-16 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Translation Inhibitor Rocaglamide Targets a Bimolecular Cavity between eIF4A and Polypurine RNA.
Mol. Cell, 73, 2019
|
|
