6J6W
 
 | | Mutant K23N of heat shock factor 4-DBD | | Descriptor: | Heat shock factor protein 4, NITRATE ION, SODIUM ION | | Authors: | Xiao, Z.Y, Cui, L.W, Wang, S, Liu, W. | | Deposit date: | 2019-01-16 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | Structure of the mutant K23N of heat shock factor 4-DBD at 1.69 Angstroms resolution.
To Be Published
|
|
4JHB
 
 | |
1FTT
 
 | | THYROID TRANSCRIPTION FACTOR 1 HOMEODOMAIN (RATTUS NORVEGICUS) | | Descriptor: | THYROID TRANSCRIPTION FACTOR 1 HOMEODOMAIN | | Authors: | Fogolari, F, Esposito, G, Damante, G, Formisano, S, Di Lauro, R, Viglino, P. | | Deposit date: | 1995-10-03 | | Release date: | 1996-01-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Analysis of the solution structure of the homeodomain of rat thyroid transcription factor 1 by 1H-NMR spectroscopy and restrained molecular mechanics.
Eur.J.Biochem., 241, 1996
|
|
4BKX
 
 | | The structure of HDAC1 in complex with the dimeric ELM2-SANT domain of MTA1 from the NuRD complex | | Descriptor: | ACETATE ION, HISTONE DEACETYLASE 1, METASTASIS-ASSOCIATED PROTEIN MTA1, ... | | Authors: | Millard, C.J, Watson, P.J, Celardo, I, Gordiyenko, Y, Cowley, S.M, Robinson, C.V, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2013-04-30 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Class I Hdacs Share a Common Mechanism of Regulation by Inositol Phosphates.
Mol.Cell, 51, 2013
|
|
2FXA
 
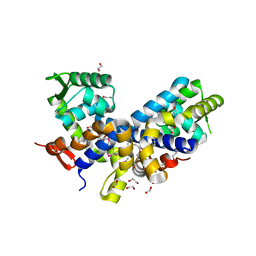 | | Structure of the Protease Production Regulatory Protein hpr from Bacillus subtilis. | | Descriptor: | 1,2-ETHANEDIOL, HEXAETHYLENE GLYCOL, PENTAETHYLENE GLYCOL, ... | | Authors: | Cuff, M.E, Skarina, T, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-03 | | Release date: | 2006-03-14 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Protease Production Regulatory Protein hpr from Bacillus subtilis.
TO BE PUBLISHED
|
|
4LNO
 
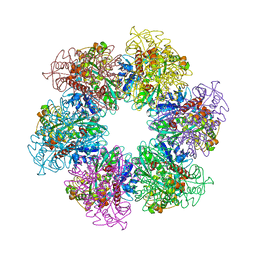 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: form two of GS-1 | | Descriptor: | GLUTAMINE, Glutamine synthetase, MAGNESIUM ION | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | Deposit date: | 2013-07-11 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
1KBH
 
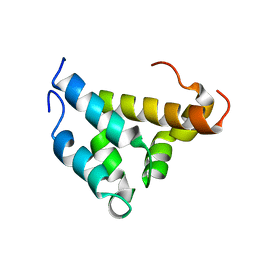 | | Mutual Synergistic Folding in the Interaction Between Nuclear Receptor Coactivators CBP and ACTR | | Descriptor: | CREB-BINDING PROTEIN, nuclear receptor coactivator | | Authors: | Demarest, S.J, Martinez-Yamout, M, Chung, J, Chen, H, Xu, W, Dyson, H.J, Evans, R.M, Wright, P.E. | | Deposit date: | 2001-11-06 | | Release date: | 2002-02-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators.
Nature, 415, 2002
|
|
6QNX
 
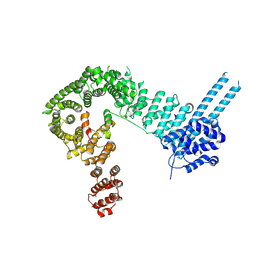 | | Structure of the SA2/SCC1/CTCF complex | | Descriptor: | Cohesin subunit SA-2, Double-strand-break repair protein rad21 homolog, Transcriptional repressor CTCF | | Authors: | Li, Y, Muir, K.W, Panne, D. | | Deposit date: | 2019-02-12 | | Release date: | 2020-01-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis for cohesin-CTCF-anchored loops.
Nature, 578, 2020
|
|
5WUQ
 
 | | Crystal structure of SigW in complex with its anti-sigma RsiW, a zinc binding form | | Descriptor: | Anti-sigma-W factor RsiW, ECF RNA polymerase sigma factor SigW, ZINC ION | | Authors: | Devkota, S.R, Kwon, E, Ha, S.C, Kim, D.Y. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the regulation of Bacillus subtilis SigW activity by anti-sigma RsiW
PLoS ONE, 12, 2017
|
|
7QH5
 
 | |
5WUR
 
 | | Crystal structure of SigW in complex with its anti-sigma RsiW, an oxdized form | | Descriptor: | Anti-sigma-W factor RsiW, ECF RNA polymerase sigma factor SigW | | Authors: | Devkota, S.R, Kwon, E, Ha, S.C, Kim, D.Y. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the regulation of Bacillus subtilis SigW activity by anti-sigma RsiW
PLoS ONE, 12, 2017
|
|
8FMU
 
 | | Crystal structure of human Brachyury G177D variant in complex with SJF-4601 | | Descriptor: | N-(3-chloro-4-fluorophenyl)-3-[4-(dimethylamino)butanamido]-4-methoxybenzamide, T-box transcription factor T | | Authors: | Bebenek, A, Linhares, B, Jaime-Figueroa, S, Butrin, A, Crews, C. | | Deposit date: | 2022-12-24 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery and Development of the First Selective Brachyury Degrader
To Be Published
|
|
6OLZ
 
 | |
6OLG
 
 | |
5ON6
 
 | | Crystal structure of haemanthamine bound to the 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Pellegrino, S, Meyer, M, Yusupova, G, Yusupov, M. | | Deposit date: | 2017-08-03 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.10000229 Å) | | Cite: | The Amaryllidaceae Alkaloid Haemanthamine Binds the Eukaryotic Ribosome to Repress Cancer Cell Growth.
Structure, 26, 2018
|
|
5OBM
 
 | | Crystal structure of Gentamicin bound to the yeast 80S ribosome | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Prokhorova, I, Djumagulov, M, Urzhumtsev, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2017-06-28 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Aminoglycoside interactions and impacts on the eukaryotic ribosome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7B5M
 
 | | Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: CUL1-RBX1-SKP1-SKP2-CKSHS1-p27~Ub~ARIH1. Transition State 2 | | Descriptor: | Cullin-1, Cyclin-dependent kinase inhibitor 1B, Cyclin-dependent kinases regulatory subunit 1, ... | | Authors: | Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2020-12-05 | | Release date: | 2021-02-17 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Nature, 590, 2021
|
|
5HDK
 
 | |
5ABA
 
 | |
7ABF
 
 | | Human pre-Bact-1 spliceosome core structure | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-07 | | Release date: | 2020-12-09 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
6FX0
 
 | |
8IR1
 
 | | human nuclear pre-60S ribosomal particle - State A | | Descriptor: | 28S rRNA, 5.8S rRMA, 5S RNA, ... | | Authors: | Zhang, Y, Gao, N. | | Deposit date: | 2023-03-17 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Visualizing the nucleoplasmic maturation of human pre-60S ribosomal particles.
Cell Res., 33, 2023
|
|
1L0O
 
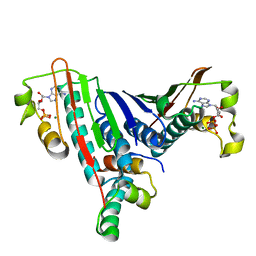 | | Crystal Structure of the Bacillus stearothermophilus Anti-Sigma Factor SpoIIAB with the Sporulation Sigma Factor SigmaF | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Anti-sigma F factor, MAGNESIUM ION, ... | | Authors: | Campbell, E.A, Masuda, S, Sun, J.L, Muzzin, O, Olson, C.A, Wang, S, Darst, S.A. | | Deposit date: | 2002-02-12 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the Bacillus stearothermophilus anti-sigma factor SpoIIAB with the sporulation sigma factor sigmaF.
Cell(Cambridge,Mass.), 108, 2002
|
|
5AOJ
 
 | | Structure of the p53 cancer mutant Y220C in complex with 2-hydroxy-3, 5-diiodo-4-(1H-pyrrol-1-yl)benzoic acid | | Descriptor: | 2-hydroxy-3,5-diiodo-4-(1H-pyrrol-1-yl)benzoic acid, CELLULAR TUMOR ANTIGEN P53, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Joerger, A.C, Baud, M.G, Bauer, M.R, Fersht, A.R. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Exploiting Transient Protein States for the Design of Small-Molecule Stabilizers of Mutant P53.
Structure, 23, 2015
|
|
5AOI
 
 | |
