3RPL
 
 | | D-fructose 1,6-bisphosphatase class 2/sedoheptulose 1,7-bisphosphatase of Synechocystis sp. PCC 6803 in complex with FRUCTOSE-1,6-BISPHOSPHATE | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Hu, X, Hui, D, Lingling, F, Jian, W. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | New insights into the structural and interactional basis for a promising route towards fructose-1,6-/sedoheptulose-1,7-bisphosphatases controlling
To be Published
|
|
5ULT
 
 | | HIV-1 wild Type protease with GRL-100-13A (a Crown-like Oxotricyclic Core as the P2-Ligand with the sulfonamide isostere as the P2' group) | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Protease, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Design and Development of Highly Potent HIV-1 Protease Inhibitors with a Crown-Like Oxotricyclic Core as the P2-Ligand To Combat Multidrug-Resistant HIV Variants.
J. Med. Chem., 60, 2017
|
|
5TKL
 
 | | Crystal structure of FBP aldolase from Toxoplasma gondii, condensation intermediate | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, 1,6-di-O-phosphono-D-fructose, Fructose-bisphosphate aldolase, ... | | Authors: | Heron, P.W, Sygusch, J. | | Deposit date: | 2016-10-06 | | Release date: | 2017-10-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Isomer activation controls stereospecificity of class I fructose-1,6-bisphosphate aldolases.
J. Biol. Chem., 292, 2017
|
|
4AG9
 
 | | C. elegans glucosamine-6-phosphate N-acetyltransferase (GNA1): ternary complex with coenzyme A and GlcNAc | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, COENZYME A, ... | | Authors: | Dorfmueller, H.C, Fang, W, Rao, F.V, Blair, D.E, Attrill, H, Shepherd, S.M, van Aalten, D.M.F. | | Deposit date: | 2012-01-24 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and Biochemical Characterization of a Trapped Coenzyme a Adduct of Caenorhabditis Elegans Glucosamine-6-Phosphate N-Acetyltransferase 1.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1ODY
 
 | | HIV-1 PROTEASE COMPLEXED WITH AN INHIBITOR LP-130 | | Descriptor: | 4-[2-(2-ACETYLAMINO-3-NAPHTALEN-1-YL-PROPIONYLAMINO)-4-METHYL-PENTANOYLAMINO]-3-HYDROXY-6-METHYL-HEPTANOIC ACID [1-(1-CARBAMOYL-2-NAPHTHALEN-1-YL-ETHYLCARBAMOYL)-PROPYL]-AMIDE, HIV-1 PROTEASE | | Authors: | Kervinen, J, Lubkowski, J, Zdanov, A, Wlodawer, A, Gustchina, A. | | Deposit date: | 1998-07-13 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Toward a universal inhibitor of retroviral proteases: comparative analysis of the interactions of LP-130 complexed with proteases from HIV-1, FIV, and EIAV.
Protein Sci., 7, 1998
|
|
3GR3
 
 | |
3EKP
 
 | | Crystal Structure of the inhibitor Amprenavir (APV) in complex with a multi-drug resistant HIV-1 protease variant (L10I/G48V/I54V/V64I/V82A)Refer: FLAP+ in citation | | Descriptor: | ACETATE ION, PHOSPHATE ION, Protease, ... | | Authors: | Prabu-Jayabalan, M, King, N.M, Bandaranayake, R.M. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Extreme Entropy-Enthalpy Compensation in a Drug-Resistant Variant of HIV-1 Protease.
Acs Chem.Biol., 7, 2012
|
|
1ON8
 
 | | Crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) with UDP and GlcUAb(1-3)Galb(1-O)-naphthalenelmethanol an acceptor substrate analog | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2, MANGANESE (II) ION, ... | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-27 | | Release date: | 2003-04-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
5UAW
 
 | | Structure of apo human PYCR-1 crystallized in space group P21212 | | Descriptor: | Pyrroline-5-carboxylate reductase 1, mitochondrial, SULFATE ION | | Authors: | Tanner, J.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Resolving the cofactor-binding site in the proline biosynthetic enzyme human pyrroline-5-carboxylate reductase 1.
J. Biol. Chem., 292, 2017
|
|
1MTB
 
 | | Viability of a drug-resistant HIV-1 protease mutant: structural insights for better antiviral therapy | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, PROTEASE RETROPEPSIN | | Authors: | Prabu-Jeyabalan, M, Nalivaika, E.A, King, N.M, Schiffer, C.A. | | Deposit date: | 2002-09-20 | | Release date: | 2003-01-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Viability of drug-resistant human immunodeficiency virus type 1 protease variant: structural insights for better antiviral therapy
J.Virol., 77, 2003
|
|
2YWP
 
 | | Crystal Structure of CHK1 with a Urea Inhibitor | | Descriptor: | 1-(5-CHLORO-2,4-DIMETHOXYPHENYL)-3-(5-CYANOPYRAZIN-2-YL)UREA, Serine/threonine-protein kinase Chk1 | | Authors: | Park, C. | | Deposit date: | 2007-04-21 | | Release date: | 2007-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synthesis and biological evaluation of 1-(2,4,5-trisubstituted phenyl)-3-(5-cyanopyrazin-2-yl)ureas as potent Chk1 kinase inhibitors
Bioorg.Med.Chem.Lett., 16, 2006
|
|
7KXW
 
 | | Crystal structure of DCLK1-KD in complex with DCLK1-IN-1 | | Descriptor: | 2-{[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}-11-methyl-5-(2,2,2-trifluoroethyl)-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, DI(HYDROXYETHYL)ETHER, Serine/threonine-protein kinase DCLK1, ... | | Authors: | Patel, O, Lucet, I. | | Deposit date: | 2020-12-05 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Structural basis for small molecule targeting of Doublecortin Like Kinase 1 with DCLK1-IN-1.
Commun Biol, 4, 2021
|
|
3EKT
 
 | | Crystal Structure of the inhibitor Darunavir (DRV) in complex with a multi-drug resistant HIV-1 protease variant (L10F/G48V/I54V/V64I/V82A) (Refer: FLAP+ in citation.) | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Prabu-Jeyabalan, M, King, N.M, Bandaranayake, R.M. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Extreme Entropy-Enthalpy Compensation in a Drug-Resistant Variant of HIV-1 Protease.
Acs Chem.Biol., 7, 2012
|
|
4IVP
 
 | | Crystal structure of thymidine kinase from herpes simplex virus type 1 in complex with IN51/20 | | Descriptor: | 6-[3-hydroxy-2-(hydroxymethyl)prop-1-en-1-yl]-5-methylpyrimidine-2,4(1H,3H)-dione, SULFATE ION, Thymidine kinase | | Authors: | Pernot, L, Novakovic, I, Westermaier, Y, Perozzo, R, Raic-Malic, S, Scapozza, L. | | Deposit date: | 2013-01-23 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New thymine derivatives as HSV1-TK ligand for the development of PET imaging tracer
To be Published
|
|
4EE3
 
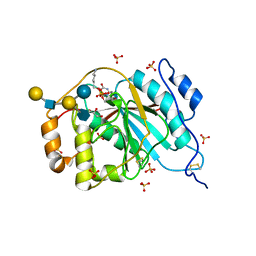 | | Crystal structure of human M340H-beta-1,4-galactosyltransferase-1 (M340H-B4GAL-T1) in complex with pentasaccharide | | Descriptor: | 6-AMINOHEXYL-URIDINE-C1,5'-DIPHOSPHATE, Beta-1,4-galactosyltransferase 1, GLYCEROL, ... | | Authors: | Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2012-03-28 | | Release date: | 2012-07-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding of N-acetylglucosamine (GlcNAc) beta 1-6-branched oligosaccharide acceptors to beta 4-galactosyltransferase I reveals a new ligand binding mode.
J.Biol.Chem., 287, 2012
|
|
5T8H
 
 | |
3F6T
 
 | |
3H5B
 
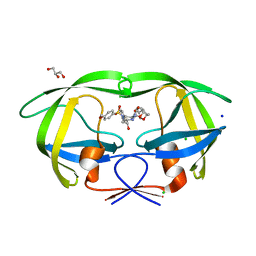 | | Crystal structure of wild type HIV-1 protease with novel P1'-ligand GRL-02031 | | Descriptor: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tie, Y, Wang, Y.F, Weber, I.T. | | Deposit date: | 2009-04-21 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Design of HIV-1 protease inhibitors with pyrrolidinones and oxazolidinones as novel P1'-ligands to enhance backbone-binding interactions with protease: synthesis, biological evaluation, and protein-ligand X-ray studies.
J.Med.Chem., 52, 2009
|
|
3HVT
 
 | | STRUCTURAL BASIS OF ASYMMETRY IN THE HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 REVERSE TRANSCRIPTASE HETERODIMER | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | Authors: | Steitz, T.A, Smerdon, S.J, Jaeger, J, Wang, J, Kohlstaedt, L.A, Chirino, A.J, Friedman, J.M, Rice, P.A. | | Deposit date: | 1994-07-25 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the binding site for nonnucleoside inhibitors of the reverse transcriptase of human immunodeficiency virus type 1.
Proc.Natl.Acad.Sci.Usa, 91, 1994
|
|
2Z4O
 
 | | Wild Type HIV-1 Protease with potent Antiviral inhibitor GRL-98065 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.F, Tie, Y, Boross, P.I, Tozser, J, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | Deposit date: | 2007-06-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease.
J.Med.Chem., 50, 2007
|
|
3EL4
 
 | | Crystal structure of inhibitor saquinavir (SQV) complexed with the multidrug HIV-1 protease variant L63P/V82T/I84V | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, ACETATE ION, Protease | | Authors: | Prabu-Jeyabalan, M, King, N, Schiffer, C. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Extreme Entropy-Enthalpy Compensation in a Drug-Resistant Variant of HIV-1 Protease.
Acs Chem.Biol., 7, 2012
|
|
1IR2
 
 | | Crystal Structure of Activated Ribulose-1,5-bisphosphate Carboxylase/oxygenase (Rubisco) from Green alga, Chlamydomonas reinhardtii Complexed with 2-Carboxyarabinitol-1,5-bisphosphate (2-CABP) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, GLYCEROL, Large subunit of Rubisco, ... | | Authors: | Mizohata, E, Matsumura, H, Okano, Y, Kumei, M, Takuma, H, Onodera, J, Kato, K, Shibata, N, Inoue, T, Yokota, A, Kai, Y. | | Deposit date: | 2001-09-03 | | Release date: | 2002-03-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of activated ribulose-1,5-bisphosphate carboxylase/oxygenase from green alga Chlamydomonas reinhardtii complexed with 2-carboxyarabinitol-1,5-bisphosphate.
J.Mol.Biol., 316, 2002
|
|
3OXC
 
 | | Wild Type HIV-1 Protease with Antiviral Drug Saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, FORMIC ACID, Protease, ... | | Authors: | Kovalevsky, A.Y, Wang, Y.-F, Tie, Y, Weber, I.T. | | Deposit date: | 2010-09-21 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir
Proteins, 67, 2007
|
|
4HYS
 
 | |
1EP4
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with S-1153 | | Descriptor: | 5-(3,5-DICHLOROPHENYL)THIO-4-ISOPROPYL-1-(PYRIDIN-4-YL-METHYL)-1H-IMIDAZOL-2-YL-METHYL CARBAMATE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Nichols, C, Bird, L.E, Fujiwara, T, Suginoto, H, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-03-27 | | Release date: | 2000-09-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding of the second generation non-nucleoside inhibitor S-1153 to HIV-1 reverse transcriptase involves extensive main chain hydrogen bonding.
J.Biol.Chem., 275, 2000
|
|
