1IIG
 
 | | STRUCTURE OF TRYPANOSOMA BRUCEI BRUCEI TRIOSEPHOSPHATE ISOMERASE COMPLEXED WITH 3-PHOSPHONOPROPIONATE | | Descriptor: | 3-PHOSPHONOPROPANOIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Noble, M.E, Wierenga, R.K, Lambeir, A.M, Opperdoes, F.R, Thunnissen, A.M, Kalk, K.H, Groendijk, H, Hol, W.G.J. | | Deposit date: | 2001-04-23 | | Release date: | 2001-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The adaptability of the active site of trypanosomal triosephosphate isomerase as observed in the crystal structures of three different complexes.
Proteins, 10, 1991
|
|
4KUH
 
 | |
6E3I
 
 | |
5P21
 
 | |
2R65
 
 | | Crystal structure of Helicobacter pylori ATP dependent protease, FtsH ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division protease ftsH homolog | | Authors: | Kim, S.H, Kang, G.B, Song, H.-E, Park, S.J, Bae, M.-H, Eom, S.H. | | Deposit date: | 2007-09-05 | | Release date: | 2008-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural studies on Helicobacter pyloriATP-dependent protease, FtsH
J.SYNCHROTRON RADIAT., 15, 2008
|
|
6E1P
 
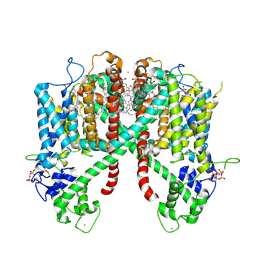 | | Structure of AtTPC1(DDE) in state 2 | | Descriptor: | CALCIUM ION, PALMITIC ACID, Two pore calcium channel protein 1 | | Authors: | Kintzer, A.F, Green, E.M, Cheng, Y, Stroud, R.M. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for activation of voltage sensor domains in an ion channel TPC1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2BWC
 
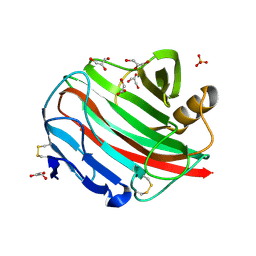 | |
2YXJ
 
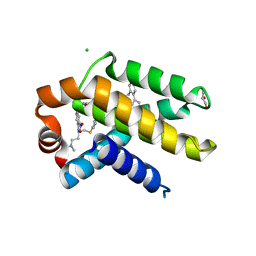 | | Crystal structure of Bcl-xL in complex with ABT-737 | | Descriptor: | 4-{4-[(4'-CHLOROBIPHENYL-2-YL)METHYL]PIPERAZIN-1-YL}-N-{[4-({(1R)-3-(DIMETHYLAMINO)-1-[(PHENYLTHIO)METHYL]PROPYL}AMINO)-3-NITROPHENYL]SULFONYL}BENZAMIDE, Apoptosis regulator Bcl-X, CHLORIDE ION, ... | | Authors: | Czabotar, P.E, Lee, E.F, Smith, B.J, Deshayes, K, Zobel, K, Fairlie, W.D, Colman, P.M. | | Deposit date: | 2007-04-26 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of ABT-737 complexed with Bcl-xL: implications for selectivity of antagonists of the Bcl-2 family
Cell Death Differ., 14, 2007
|
|
3H1L
 
 | | Chicken cytochrome BC1 complex with ascochlorin bound at QO and QI sites | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 3-chloro-4,6-dihydroxy-2-methyl-5-{(2E,4E)-3-methyl-5-[(1R,2R,6R)-1,2,6-trimethyl-3-oxocyclohexyl]penta-2,4-dien-1-yl}benzaldehyde, CARDIOLIPIN, ... | | Authors: | Berry, E.A, Huang, L.S, Minagawa, N. | | Deposit date: | 2009-04-12 | | Release date: | 2010-01-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Ascochlorin is a novel, specific inhibitor of the mitochondrial cytochrome bc(1) complex.
Biochim.Biophys.Acta, 1797, 2010
|
|
6EF0
 
 | | Yeast 26S proteasome bound to ubiquitinated substrate (1D* motor state) | | Descriptor: | 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6B homolog, ... | | Authors: | de la Pena, A.H, Goodall, E.A, Gates, S.N, Lander, G.C, Martin, A. | | Deposit date: | 2018-08-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Substrate-engaged 26Sproteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation.
Science, 362, 2018
|
|
3E6K
 
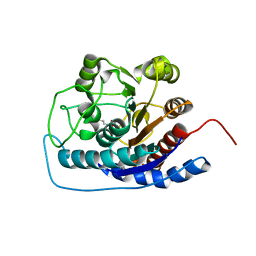 | |
3E6V
 
 | |
6EHA
 
 | | Heme oxygenase 1 in complex with inhibitor | | Descriptor: | 1-(3-imidazol-1-ylpropyl)-5-(2-methylpropyl)-4-phenyl-imidazole, Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Grudnik, P, Mieczkowski, M. | | Deposit date: | 2017-09-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development and characterization of a new inhibitor of heme oxygenase activity for cancer treatment.
Arch.Biochem.Biophys., 671, 2019
|
|
4P2C
 
 | | Complex of Shiga toxin 2e with a neutralizing single-domain antibody | | Descriptor: | Nanobody 1, Anti-F4+ETEC bacteria VHH variable region, Shiga toxin 2e, ... | | Authors: | Lo, A.W.H, Moonens, K, De Kerpel, M, Brys, L, Pardon, E, Remaut, H, De Greve, H. | | Deposit date: | 2014-03-03 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.818 Å) | | Cite: | The Molecular Mechanism of Shiga Toxin Stx2e Neutralization by a Single-domain Antibody Targeting the Cell Receptor-binding Domain.
J.Biol.Chem., 289, 2014
|
|
2LYM
 
 | |
4P2S
 
 | | Alanine Scanning Mutagenesis Identifies an Asparagine-Arginine-Lysine Triad Essential to Assembly of the Shell of the Pdu Microcompartment | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Putative propanediol utilization protein PduA, ... | | Authors: | Sinha, S, Cheng, S, Sung, Y.W, McNamara, D.E, Sawaya, M.R, Yeates, T.O, Bobik, T.A. | | Deposit date: | 2014-03-03 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Alanine Scanning Mutagenesis Identifies an Asparagine-Arginine-Lysine Triad Essential to Assembly of the Shell of the Pdu Microcompartment.
J.Mol.Biol., 426, 2014
|
|
8P45
 
 | | Crystal structure of human STING in complex with the agonist MD1202D | | Descriptor: | 9-[(1~{S},3~{R},6~{R},8~{R},9~{R},10~{R},12~{R},15~{R},17~{R},18~{R})-8-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Klima, M, Boura, E. | | Deposit date: | 2023-05-19 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Fluorinated cGAMP analogs, which act as STING agonists and are not cleavable by poxins: Structural basis of their function.
Structure, 32, 2024
|
|
4KUK
 
 | | A superfast recovering full-length LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae (Dark state) | | Descriptor: | ACETIC ACID, RIBOFLAVIN, blue-light photoreceptor | | Authors: | Circolone, F, Granzin, J, Stadler, A, Krauss, U, Drepper, T, Endres, S, Knieps-Gruenhagen, E, Wirtz, A, Willbold, D, Batra-Safferling, R, Jaeger, K.-E. | | Deposit date: | 2013-05-22 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of a short LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae.
BMC Microbiol, 15, 2015
|
|
4KUX
 
 | | Crystal structure of Aspergillus terreus aristolochene synthase complexed with farnesyl thiolodiphosphate (FSPP) | | Descriptor: | Aristolochene synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chen, M, Faraldos, J.A, Al-lami, N, Janvier, M, D'Antonio, E.L, Cane, D.E, Allemann, R.K, Christianson, D.W. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights from the binding of substrate and carbocation intermediate analogues to aristolochene synthase.
Biochemistry, 52, 2013
|
|
2RBC
 
 | | Crystal structure of a putative ribokinase from Agrobacterium tumefaciens | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Cuff, M.E, Xu, X, Zheng, H, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-18 | | Release date: | 2007-11-13 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a putative ribokinase from Agrobacterium tumefaciens.
TO BE PUBLISHED
|
|
8ORV
 
 | | Crystal structure of monkeypox virus poxin in complex with the STING agonist MD1203 | | Descriptor: | 9-[(1~{S},6~{R},8~{R},9~{R},10~{R},15~{R},17~{R},18~{R})-8-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, MPXVgp165 | | Authors: | Duchoslav, V, Klima, M, Boura, E. | | Deposit date: | 2023-04-17 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Fluorinated cGAMP analogs, which act as STING agonists and are not cleavable by poxins: Structural basis of their function.
Structure, 32, 2024
|
|
4ZNH
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with a 2-Fluoro-substituted OBHS derivative | | Descriptor: | 2-fluorophenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor-interacting peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-04 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
8ORW
 
 | | Crystal structure of human STING in complex with the agonist MD1203 | | Descriptor: | 9-[(1~{S},6~{R},8~{R},9~{R},10~{R},15~{R},17~{R},18~{R})-8-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon protein | | Authors: | Klima, M, Boura, E. | | Deposit date: | 2023-04-17 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Fluorinated cGAMP analogs, which act as STING agonists and are not cleavable by poxins: Structural basis of their function.
Structure, 32, 2024
|
|
5OW5
 
 | | p60p80-CAMSAP complex | | Descriptor: | 1,2-ETHANEDIOL, Calmodulin-regulated spectrin-associated protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rezabkova, L, Capitani, G, Prota, A.E, Kammerer, R.A, Steinmetz, M.O. | | Deposit date: | 2017-08-30 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Formation of the Microtubule Minus-End-Regulating CAMSAP-Katanin Complex.
Structure, 26, 2018
|
|
4ZNT
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with a 3-Bromo-substituted OBHS derivative | | Descriptor: | 3-bromophenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor-interacting peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-05 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
