3BCG
 
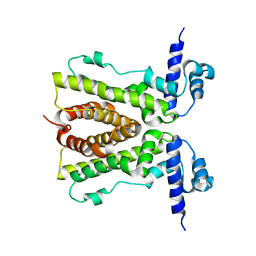 | | Conformational changes of the AcrR regulator reveal a mechanism of induction | | Descriptor: | HTH-type transcriptional regulator acrR | | Authors: | Gu, R, Li, M, Su, C.C, Long, F, Yang, F, McDermott, G, Yu, E.Y. | | Deposit date: | 2007-11-12 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Conformational change of the AcrR regulator reveals a possible mechanism of induction.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
4WWW
 
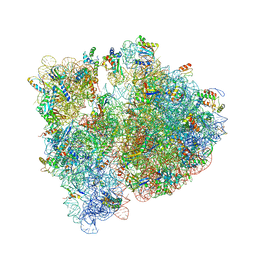 | | Crystal structure of the E. coli ribosome bound to CEM-101 | | Descriptor: | (3aS,4R,7S,9R,10R,11R,13R,15R,15aR)-1-{4-[4-(3-aminophenyl)-1H-1,2,3-triazol-1-yl]butyl}-4-ethyl-7-fluoro-11-methoxy-3a ,7,9,11,13,15-hexamethyl-2,6,8,14-tetraoxotetradecahydro-2H-oxacyclotetradecino[4,3-d][1,3]oxazol-10-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 16S rRNA, 23S rRNA, ... | | Authors: | Dunkle, J.A, Zhang, W, Cate, J.H.D, Mankin, A.S. | | Deposit date: | 2014-11-12 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Binding and action of CEM-101, a new fluoroketolide antibiotic that inhibits protein synthesis.
Antimicrob. Agents Chemother., 54, 2010
|
|
5LN3
 
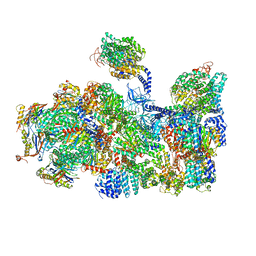 | | The human 26S Proteasome at 6.8 Ang. | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Schweitzer, A, Beck, F, Sakata, E, Unverdorben, P. | | Deposit date: | 2016-08-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Molecular Details Underlying Dynamic Structures and Regulation of the Human 26S Proteasome.
Mol. Cell Proteomics, 16, 2017
|
|
4WF1
 
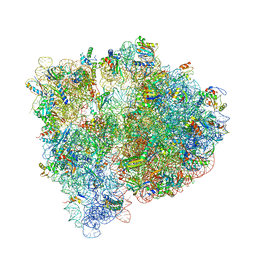 | | Crystal structure of the E. coli ribosome bound to negamycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Olivier, N.B, Altman, R.B, Noeske, J, Basarab, G.S, Code, E, Ferguson, A.D, Gao, N, Huang, J, Juette, M.F, Livchak, S, Miller, M.D, Prince, D.B, Cate, J.H.D, Buurman, E.T, Blanchard, S.C. | | Deposit date: | 2014-09-11 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Negamycin induces translational stalling and miscoding by binding to the small subunit head domain of the Escherichia coli ribosome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8I0T
 
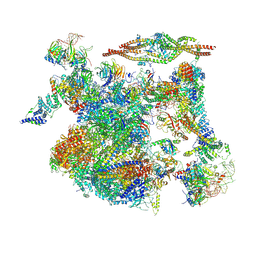 | | The cryo-EM structure of human Bact-III complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Crooked neck-like protein 1, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0V
 
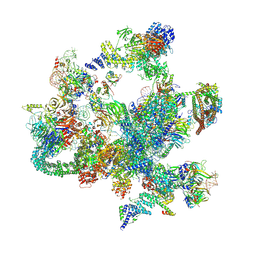 | | The cryo-EM structure of human post-Bact complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Crooked neck-like protein 1, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0R
 
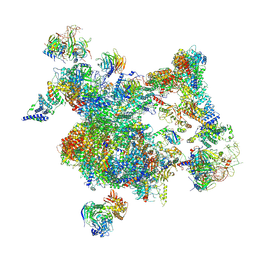 | | The cryo-EM structure of human Bact-I complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0S
 
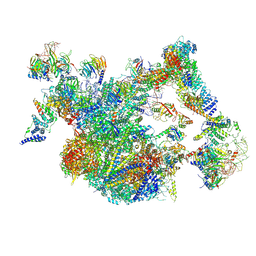 | | The cryo-EM structure of human Bact-II complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Crooked neck-like protein 1, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0W
 
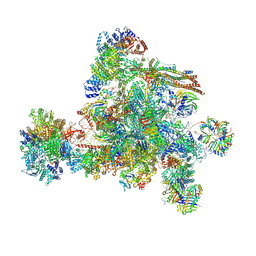 | | The cryo-EM structure of human C complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Coiled-coil domain-containing protein 94, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0U
 
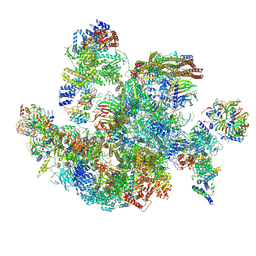 | | The cryo-EM structure of human Bact-IV complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Crooked neck-like protein 1, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8EIU
 
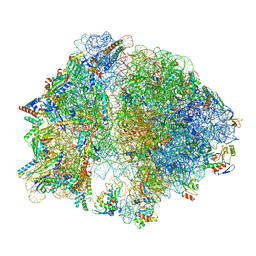 | | E. coli 70S ribosome with A-loop mutations U2554C and U2555C | | Descriptor: | 16S rRNA, 23S rRNA, 3'-amino-3'-deoxyadenosine 5'-(dihydrogen phosphate), ... | | Authors: | Nissley, A.J, Penev, P.I, Watson, Z.L, Banfield, J.F, Cate, J.H.D. | | Deposit date: | 2022-09-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Rare ribosomal RNA sequences from archaea stabilize the bacterial ribosome.
Nucleic Acids Res., 51, 2023
|
|
7UXH
 
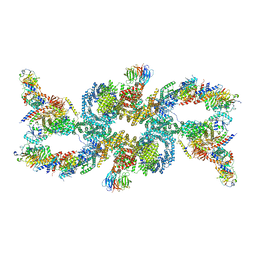 | | cryo-EM structure of the mTORC1-TFEB-Rag-Ragulator complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Cui, Z, Hurley, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the lysosomal mTORC1-TFEB-Rag-Ragulator megacomplex.
Nature, 614, 2023
|
|
7UXC
 
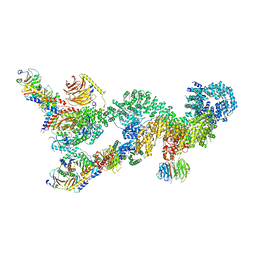 | |
8EMM
 
 | |
8CH6
 
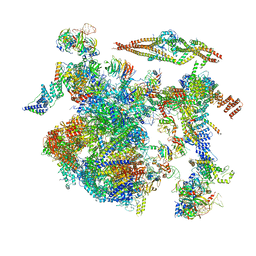 | | Structure of a late-stage activated spliceosome (BAqr) arrested with a dominant-negative Aquarius mutant (state B complex). | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Cretu, C, Schmitzova, J, Pena, V. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of catalytic activation in human splicing.
Nature, 617, 2023
|
|
1JH9
 
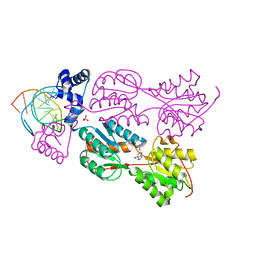 | | PURINE REPRESSOR MUTANT-HYPOXANTHINE-PURF OPERATOR COMPLEX | | Descriptor: | 5'-D(*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*TP*TP*TP*TP*CP*GP*T)-3', HYPOXANTHINE, PHOSPHATE ION, ... | | Authors: | Huffman, J.L, Lu, F, Zalkin, H, Brennan, R.G. | | Deposit date: | 2001-06-27 | | Release date: | 2002-02-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Role of residue 147 in the gene regulatory function of the Escherichia coli purine repressor.
Biochemistry, 41, 2002
|
|
8RGL
 
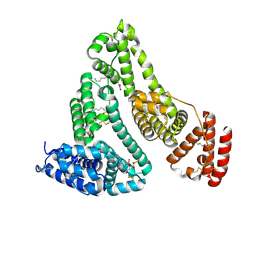 | | Structure of Human Serum Albumin in complex with Aristolochic Acid I at 1.9 A resolution - Optimized | | Descriptor: | 1,2-ETHANEDIOL, 8-methoxy-6-nitro-naphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, MYRISTIC ACID, ... | | Authors: | Pomyalov, S, Sidorenko, V.S, Grollman, A.P, Shoham, G. | | Deposit date: | 2023-12-13 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mechanistic insights into the transport of aristolochic acids and their active metabolites by human serum albumin.
J.Biol.Chem., 300, 2024
|
|
6WDF
 
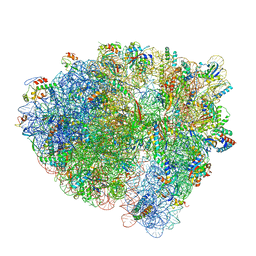 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure VI-A) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
8RGK
 
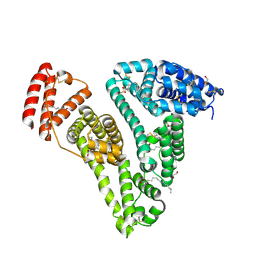 | | Structure of Human Serum Albumin in complex with Aristolochic Acid at 1.9 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 8-methoxy-6-nitro-naphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, MYRISTIC ACID, ... | | Authors: | Pomyalov, S, Sidorenko, V.S, Grollman, A.P, Shoham, G. | | Deposit date: | 2023-12-13 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mechanistic insights into the transport of aristolochic acids and their active metabolites by human serum albumin.
J.Biol.Chem., 300, 2024
|
|
8RCO
 
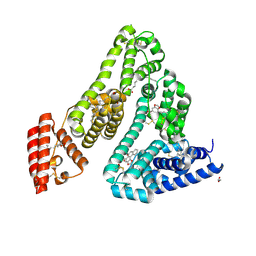 | | Structure of Human Serum Albumin in complex with Aristolochic Acid II at 1.9 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 6-nitronaphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, MYRISTIC ACID, ... | | Authors: | Pomyalov, S, Sidorenko, V.S, Grollman, A.P, Shoham, G. | | Deposit date: | 2023-12-06 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mechanistic insights into the transport of aristolochic acids and their active metabolites by human serum albumin.
J.Biol.Chem., 300, 2024
|
|
8RCP
 
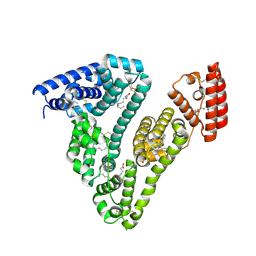 | | Structure of Human Serum Albumin in complex with Myristic Acid | | Descriptor: | 1,2-ETHANEDIOL, MYRISTIC ACID, Serum albumin | | Authors: | Pomyalov, S, Sidorenko, V.S, Grollman, A.P, Shoham, G. | | Deposit date: | 2023-12-06 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mechanistic insights into the transport of aristolochic acids and their active metabolites by human serum albumin.
J.Biol.Chem., 300, 2024
|
|
6WDJ
 
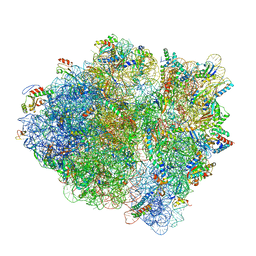 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Non-cognate Structure V-A1) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
7NTF
 
 | | Cryo-EM structure of unliganded O-GlcNAc transferase | | Descriptor: | Isoform 1 of UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Meek, R.W, Blaza, J.N, Davies, G.J. | | Deposit date: | 2021-03-09 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5.32 Å) | | Cite: | Cryo-EM structure provides insights into the dimer arrangement of the O-linked beta-N-acetylglucosamine transferase OGT.
Nat Commun, 12, 2021
|
|
7NUD
 
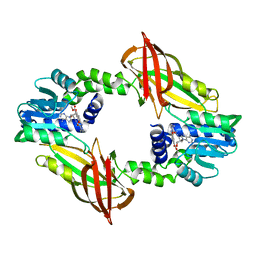 | | Crystal structure of mouse PRMT6 in complex with inhibitor EML734 | | Descriptor: | Protein arginine N-methyltransferase 6, methyl 6-[3-[[~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]carbamimidoyl]amino]propylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
7NUE
 
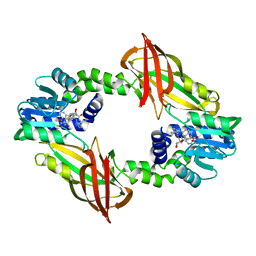 | | Crystal structure of mouse PRMT6 in complex with inhibitor EML736 | | Descriptor: | Protein arginine N-methyltransferase 6, methyl 6-[5-[[~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]carbamimidoyl]amino]pentylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
