8F0W
 
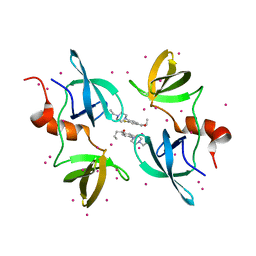 | | Tudor Domain of Tumor suppressor p53BP1 with MFP-5956 | | Descriptor: | 1-[4-(4-ethylpiperazin-1-yl)-3-fluorophenyl]butan-1-one, TP53-binding protein 1, UNKNOWN ATOM OR ION | | Authors: | The, J, Hong, Z, Dong, A, Headey, S, Gunzburg, M, Doak, B, James, L.I, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-04 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Tudor Domain of Tumor suppressor p53BP1 with MFP-5956
to be published
|
|
8EQH
 
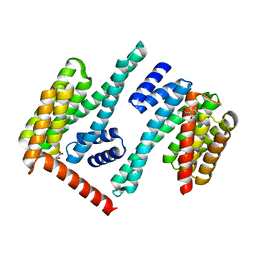 | |
5TLZ
 
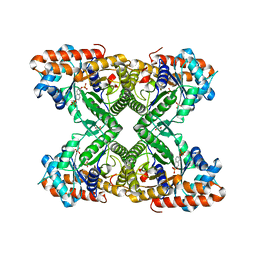 | | Fructose-1,6-bisphosphate aldolase from rabbit muscle in complex with the inhibitor naphthalene 2,6-bisphosphate | | Descriptor: | Fructose-bisphosphate aldolase A, GLYCEROL, naphthalene-2,6-diyl bis[dihydrogen (phosphate)] | | Authors: | Heron, P.W, Sygusch, J. | | Deposit date: | 2016-10-12 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Bisphosphonate Inhibitors of Mammalian Glycolytic Aldolase.
J.Med.Chem., 61, 2018
|
|
3B97
 
 | | Crystal Structure of human Enolase 1 | | Descriptor: | Alpha-enolase, MAGNESIUM ION, SULFATE ION | | Authors: | Kang, H.J, Jung, S.K, Kim, S.J, Chung, S.J. | | Deposit date: | 2007-11-02 | | Release date: | 2008-09-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human alpha-enolase (hENO1), a multifunctional glycolytic enzyme.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
5TA8
 
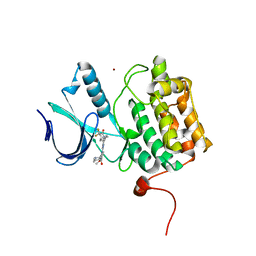 | | Crystal structure of PLK1 in complex with a novel 5,6-dihydroimidazolo[1,5-f]pteridine inhibitor | | Descriptor: | 4-[(9-cyclopentyl-7,7-difluoro-5-methyl-6-oxo-6,7,8,9-tetrahydro-5H-pyrimido[4,5-b][1,4]diazepin-2-yl)amino]-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Serine/threonine-protein kinase PLK1, ZINC ION | | Authors: | Skene, R.J, Hosfield, D.J. | | Deposit date: | 2016-09-09 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design and SAR development of 5,6-dihydroimidazolo[1,5-f]pteridine derivatives as novel Polo-like kinase-1 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5TC4
 
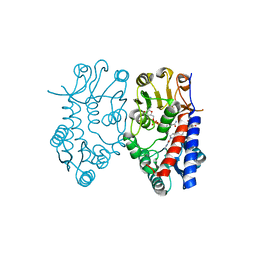 | | Crystal structure of human mitochondrial methylenetetrahydrofolate dehydrogenase-cyclohydrolase (MTHFD2) in complex with LY345899 and cofactors | | Descriptor: | 4-(7-AMINO-9-HYDROXY-1-OXO-3,3A,4,5-TETRAHYDRO-2,5,6,8,9B-PENTAAZA-CYCLOPENTA[A]NAPHTHALEN-2-YL)-PHENYLCARBONYL-GLUTAMI C ACID, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, ... | | Authors: | Gustafsson, R, Jemth, A.-S, Gustafsson Sheppard, N, Farnegardh, K, Loseva, O, Wiita, E, Bonagas, N, Dahllund, L, Llona-Minguez, S, Haggblad, M, Henriksson, M, Andersson, Y, Homan, E, Helleday, T, Stenmark, P. | | Deposit date: | 2016-09-14 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of the Emerging Cancer Target MTHFD2 in Complex with a Substrate-Based Inhibitor.
Cancer Res., 77, 2017
|
|
3B8D
 
 | | Fructose 1,6-bisphosphate aldolase from rabbit muscle | | Descriptor: | Fructose-bisphosphate aldolase A, SULFATE ION | | Authors: | Maurady, A, Sygusch, J. | | Deposit date: | 2007-11-01 | | Release date: | 2007-11-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A conserved glutamate residue exhibits multifunctional catalytic roles in D-fructose-1,6-bisphosphate aldolases.
J.Biol.Chem., 277, 2002
|
|
3BDL
 
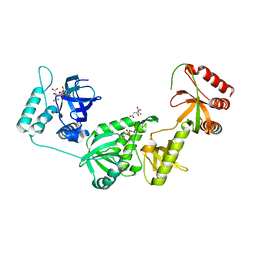 | | Crystal structure of a truncated human Tudor-SN | | Descriptor: | CITRIC ACID, Staphylococcal nuclease domain-containing protein 1 | | Authors: | Li, C.L. | | Deposit date: | 2007-11-15 | | Release date: | 2008-08-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insights into human Tudor-SN, a key component linking RNA interference and editing.
Nucleic Acids Res., 36, 2008
|
|
5UWT
 
 | |
3BI7
 
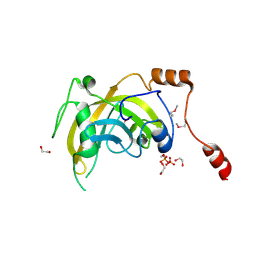 | | Crystal structure of the SRA domain of E3 ubiquitin-protein ligase UHRF1 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1, SULFATE ION, ... | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1.
Nature, 455, 2008
|
|
5UVW
 
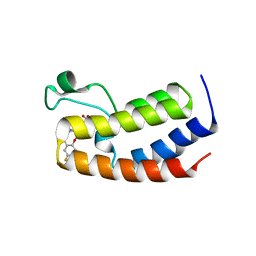 | | BRD4_Bromodomain1-A1376855 | | Descriptor: | Bromodomain-containing protein 4, N-[4-(2,4-difluorophenoxy)-3-(6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)phenyl]ethanesulfonamide, SULFATE ION | | Authors: | Park, C.H. | | Deposit date: | 2017-02-20 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | BRD4_Bromodomain1-A1376855
To Be Published
|
|
5UWS
 
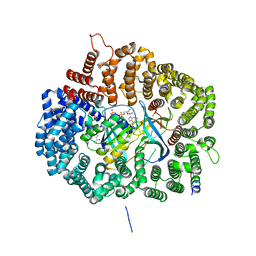 | |
5V9T
 
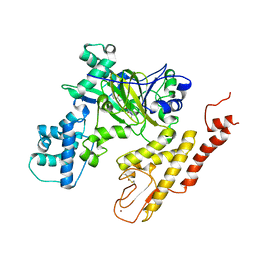 | | Crystal structure of selective pyrrolidine amide KDM5a inhibitor N-{(3R)-1-[3-(propan-2-yl)-1H-pyrazole-5-carbonyl]pyrrolidin-3-yl}cyclopropanecarboxamide (compound 48) | | Descriptor: | Lysine-specific demethylase 5A, N-{(3R)-1-[3-(propan-2-yl)-1H-pyrazole-5-carbonyl]pyrrolidin-3-yl}cyclopropanecarboxamide, NICKEL (II) ION, ... | | Authors: | Kiefer, J.R, Liang, J, Vinogradova, M. | | Deposit date: | 2017-03-23 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | From a novel HTS hit to potent, selective, and orally bioavailable KDM5 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5VBP
 
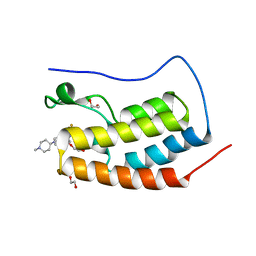 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH RO3280 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(9-cyclopentyl-7,7-difluoro-5-methyl-6-oxo-6,7,8,9-tetrahydro-5H-pyrimido[4,5-b][1,4]diazepin-2-yl)amino]-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Bromodomain-containing protein 4, ... | | Authors: | EMBER, S.W, ZHU, J.-Y, SCHONBRUNN, E. | | Deposit date: | 2017-03-30 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 2021
|
|
5UPD
 
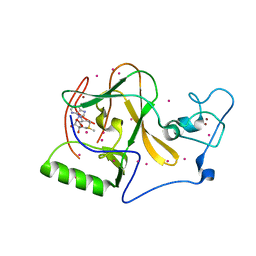 | | Methyltransferase domain of human Wolf-Hirschhorn Syndrome Candidate 1-Like protein 1 (WHSC1L1) | | Descriptor: | Histone-lysine N-methyltransferase NSD3, S-ADENOSYLMETHIONINE, UNKNOWN ATOM OR ION, ... | | Authors: | Tempel, W, Yu, W, Dong, A, Cerovina, T, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-02-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Methyltransferase domain of human Wolf-Hirschhorn Syndrome Candidate 1-Like protein 1 (WHSC1L1)
To Be Published
|
|
2YEL
 
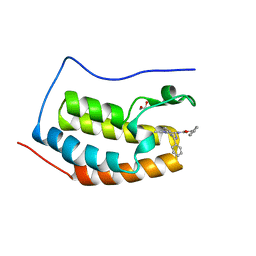 | | Crystal Structure of the First Bromodomain of Human Brd4 with the inhibitor GW841819X | | Descriptor: | 1,2-ETHANEDIOL, BENZYL [(4R)-1-METHYL-6-PHENYL-4H-[1,2,4]TRIAZOLO[4,3-A][1,4]BENZODIAZEPIN-4-YL]CARBAMATE, HUMAN BRD4 | | Authors: | Chung, C.W. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and Characterization of Small Molecule Inhibitors of the Bet Family Bromodomains.
J.Med.Chem., 54, 2011
|
|
5VDC
 
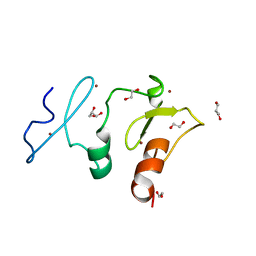 | | Crystal structure of the human DPF2 tandem PHD finger domain | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, ZINC ION, ... | | Authors: | Huber, F.M, Davenport, A.M, Hoelz, A. | | Deposit date: | 2017-04-01 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Histone-binding of DPF2 mediates its repressive role in myeloid differentiation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VNA
 
 | | Crystal structure of human YEATS domain | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, SULFATE ION, ... | | Authors: | Cho, H.J, Cierpicki, T. | | Deposit date: | 2017-04-29 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | GAS41 Recognizes Diacetylated Histone H3 through a Bivalent Binding Mode.
ACS Chem. Biol., 13, 2018
|
|
2ZO2
 
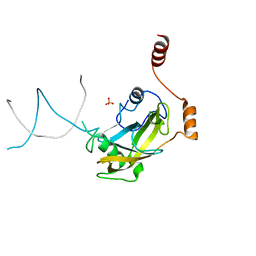 | | Mouse NP95 SRA domain non-specific DNA complex | | Descriptor: | DNA (5'-D(*DAP*DAP*DCP*DTP*DGP*DCP*DGP*DCP*DAP*DGP*DTP*DT)-3'), E3 ubiquitin-protein ligase UHRF1, PHOSPHATE ION | | Authors: | Hashimoto, H, Horton, J.R, Cheng, X. | | Deposit date: | 2008-05-05 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix
Nature, 455, 2008
|
|
2Z5U
 
 | |
3AV5
 
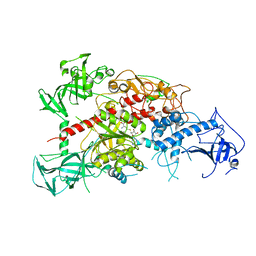 | | Crystal structure of mouse DNA methyltransferase 1 with AdoHcy | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Takeshita, K, Suetake, I, Yamashita, E, Suga, M, Narita, H, Nakagawa, A, Tajima, S. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5UWP
 
 | |
3AV6
 
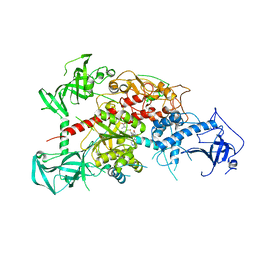 | | Crystal structure of mouse DNA methyltransferase 1 with AdoMet | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | Takeshita, K, Suetake, I, Yamashita, E, Suga, M, Narita, H, Nakagawa, A, Tajima, S. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5V67
 
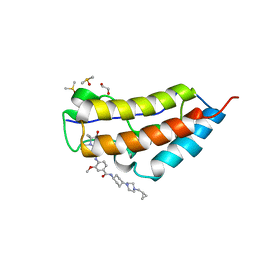 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH Volasertib | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, ... | | Authors: | EMBER, S.W, ZHU, J.-Y, SCHONBRUNN, E. | | Deposit date: | 2017-03-16 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 2021
|
|
5UWO
 
 | |
