8FQ3
 
 | | LBD conformation 2 (LBDconf2) of GluA2 flip Q isoform N619K mutant of AMPA receptor in complex with gain-of-function TARP gamma-2, with 10mM CaCl2, 150mM NaCl, 1mM MgCl2, 330uM CTZ, and 100mM glutamate (Open-CaNaMg/N619K) | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2 | | Authors: | Nakagawa, T. | | Deposit date: | 2023-01-05 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | The open gate of the AMPA receptor forms a Ca 2+ binding site critical in regulating ion transport.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FPC
 
 | | Conformation 1 of the ligand binding domain (LBDconf1) of GluA2 flip Q isoform of AMPA receptor in complex with gain-of-function TARP gamma-2, with 10mM CaCl2, 150mM NaCl, 1mM MgCl2, 330uM CTZ, and 100mM glutamate (Open-CaNaMg) | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2 | | Authors: | Nakagawa, T. | | Deposit date: | 2023-01-04 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | The open gate of the AMPA receptor forms a Ca 2+ binding site critical in regulating ion transport.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FPH
 
 | | Conformation 2 of the ligand binding domain (LBDconf2) of GluA2 flip Q isoform of AMPA receptor in complex with gain-of-function TARP gamma-2, with 10mM CaCl2, 150mM NaCl, 1mM MgCl2, 330uM CTZ, and 100mM glutamate (Open-CaNaMg) | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2 | | Authors: | Nakagawa, T. | | Deposit date: | 2023-01-04 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The open gate of the AMPA receptor forms a Ca 2+ binding site critical in regulating ion transport.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FQ8
 
 | |
8FQD
 
 | |
8FPL
 
 | | LBD conformation 2 (LBDconf2) of GluA2 flip Q isoform of AMPA receptor in complex with gain-of-function TARP gamma-2, with 10mM CaCl2, 150mM NaCl, 1mM MgCl2, 330uM CTZ, and 100uM CNQX (Closed-CaNaMg) | | Descriptor: | 6-(aminomethyl)-7-nitro-1,4-dihydroquinoxaline-2,3-dione, CYCLOTHIAZIDE, Glutamate receptor 2 | | Authors: | Nakagawa, T. | | Deposit date: | 2023-01-04 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | The open gate of the AMPA receptor forms a Ca 2+ binding site critical in regulating ion transport.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FPK
 
 | | LBD conformation 1 of GluA2 flip Q isoform of AMPA receptor in complex with gain-of-function TARP gamma-2, with 10mM CaCl2, 150mM NaCl, 1mM MgCl2, 330uM CTZ, and 100uM CNQX (Closed-CaNaMg) | | Descriptor: | 6-(aminomethyl)-7-nitro-1,4-dihydroquinoxaline-2,3-dione, CYCLOTHIAZIDE, Glutamate receptor 2 | | Authors: | Nakagawa, T. | | Deposit date: | 2023-01-04 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | The open gate of the AMPA receptor forms a Ca 2+ binding site critical in regulating ion transport.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FQA
 
 | |
8FQ2
 
 | | LBD conformation 1 (LBDconf1) of GluA2 flip Q isoform N619K mutant of AMPA receptor in complex with gain-of-function TARP gamma-2, with 10mM CaCl2, 150mM NaCl, 1mM MgCl2, 330uM CTZ, and 100mM glutamate (Open-CaNaMg/N619K) | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2 | | Authors: | Nakagawa, T. | | Deposit date: | 2023-01-05 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The open gate of the AMPA receptor forms a Ca 2+ binding site critical in regulating ion transport.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SS8
 
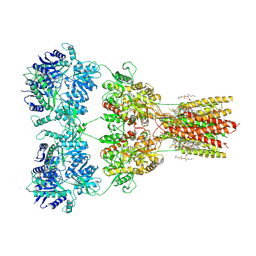 | | Structure of AMPA receptor GluA2 complex with auxiliary subunit TARP gamma-5 bound to competitive antagonist ZK and antiepileptic drug perampanel (closed state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(6'-oxo-1'-phenyl[1',6'-dihydro[2,3'-bipyridine]]-5'-yl)benzonitrile, CHOLESTEROL, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8SS7
 
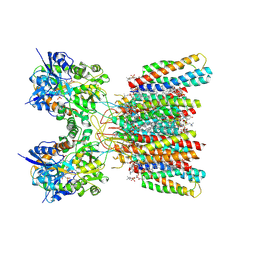 | | Structure of AMPA receptor GluA2 complex with auxiliary subunits TARP gamma-5 and cornichon-2 bound to competitive antagonist ZK, channel blocker spermidine and antiepileptic drug perampanel (closed state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(6'-oxo-1'-phenyl[1',6'-dihydro[2,3'-bipyridine]]-5'-yl)benzonitrile, CHOLESTEROL, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8SS9
 
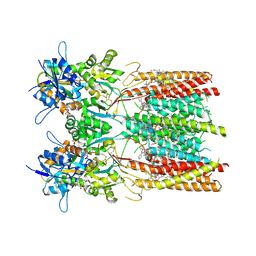 | | Structure of LBD-TMD of AMPA receptor GluA2 in complex with auxiliary subunit TARP gamma-5 bound to competitive antagonist ZK and antiepileptic drug perampanel (closed state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(6'-oxo-1'-phenyl[1',6'-dihydro[2,3'-bipyridine]]-5'-yl)benzonitrile, CHOLESTEROL, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8SS6
 
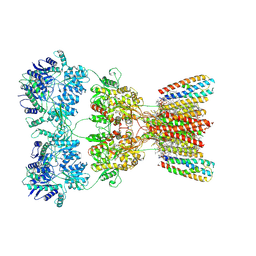 | | Structure of AMPA receptor GluA2 complex with auxiliary subunits TARP gamma-5 and cornichon-2 bound to competitive antagonist ZK, channel blocker spermidine and antiepileptic drug perampanel (closed state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(6'-oxo-1'-phenyl[1',6'-dihydro[2,3'-bipyridine]]-5'-yl)benzonitrile, CHOLESTEROL, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8YH4
 
 | | Chito oligosaccharide deacetylase from vibrio campbellii (VhCOD) in complex with chitosanbiose (GlcN)2 | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, NodB homology domain-containing protein, SODIUM ION, ... | | Authors: | Sirikan, P, Tamo, F, Robinson, R.C, Wipa, S. | | Deposit date: | 2024-02-27 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.128 Å) | | Cite: | Chito oligosaccharide deacetylase from vibrio campbellii (VhCOD) in complex with chitosanbiose (GlcN)2
To be published
|
|
8FWV
 
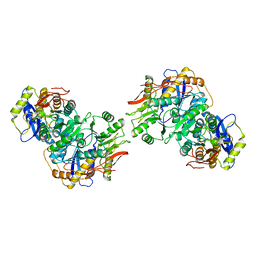 | | Structure of the amino-terminal domain of kainate receptor GluK2 in complex with the positive allosteric modulator BPAM344 and noncompetitive inhibitor perampanel | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-01-23 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Positive and negative allosteric modulation of GluK2 kainate receptors by BPAM344 and antiepileptic perampanel.
Cell Rep, 42, 2023
|
|
8FWW
 
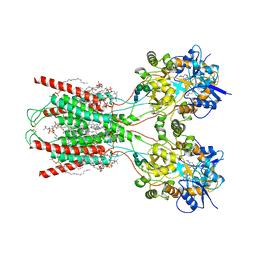 | | Structure of the ligand-binding and transmembrane domains of kainate receptor GluK2 in complex with the positive allosteric modulator BPAM344 and noncompetitive inhibitor perampanel | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-(6'-oxo-1'-phenyl[1',6'-dihydro[2,3'-bipyridine]]-5'-yl)benzonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-01-23 | | Release date: | 2023-04-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Positive and negative allosteric modulation of GluK2 kainate receptors by BPAM344 and antiepileptic perampanel.
Cell Rep, 42, 2023
|
|
6WGW
 
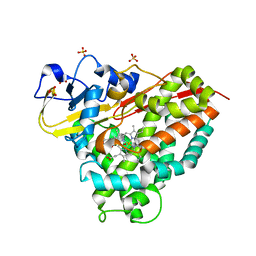 | | CYP101D1 D259E Hydroxycamphor bound | | Descriptor: | 5-EXO-HYDROXYCAMPHOR, CAMPHOR, Cytochrome P450 101D1, ... | | Authors: | Amaya, J.A, Poulos, T.L, Batabyal, D. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Proton Relay Network in the Bacterial P450s: CYP101A1 and CYP101D1.
Biochemistry, 59, 2020
|
|
9JA8
 
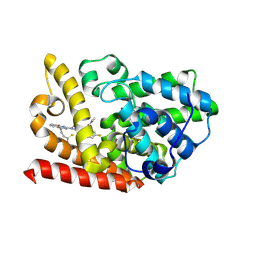 | | Crystal structure of human phosphodiesterase 10A in complex with N-(2-amino-2-thioxoethyl)-2-(3-(3-(dimethylcarbamoyl)-6-fluoroimidazo[1,2-a]pyridin-2-yl)azetidin-1-yl)quinoline-4-carboxamide | | Descriptor: | MAGNESIUM ION, ZINC ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A, ... | | Authors: | Zhang, F.C, Huang, Y.Y, Luo, H.B, Guo, L. | | Deposit date: | 2024-08-24 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhalable Carbonyl Sulfide Donor-Hybridized Selective Phosphodiesterase 10A Inhibitor for Treating Idiopathic Pulmonary Fibrosis by Inhibiting Tumor Growth Factor-beta Signaling and Activating the cAMP/Protein Kinase A/cAMP Response Element-Binding Protein (CREB)/p53 Axis.
Acs Pharmacol Transl Sci, 8, 2025
|
|
8FQE
 
 | |
9JNN
 
 | | Structure of native di-heteromeric GluN1-GluN2B NMDA receptor in rat cortex and hippocampus | | Descriptor: | (2R)-4-(3-phosphonopropyl)piperazine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, M, Feng, J, Li, Y, Zhu, S. | | Deposit date: | 2024-09-23 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Assembly and architecture of endogenous NMDA receptors in adult cerebral cortex and hippocampus.
Cell, 188, 2025
|
|
4TSZ
 
 | | Crystal structure of DNA polymerase sliding clamp from Pseudomonas aeruginosa with ligand | | Descriptor: | ACE-GLN-ALC-ASP-LEU-ZCL peptide, DNA polymerase III subunit beta | | Authors: | Olieric, V, Burnouf, D, Ennifar, E, Wolff, P. | | Deposit date: | 2014-06-19 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
1JQJ
 
 | | Mechanism of Processivity Clamp Opening by the Delta Subunit Wrench of the Clamp Loader Complex of E. coli DNA Polymerase III: Structure of the beta-delta complex | | Descriptor: | DNA polymerase III, beta chain, delta subunit | | Authors: | Jeruzalmi, D, Yurieva, O, Zhao, Y, Young, M, Stewart, J, Hingorani, M, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2001-08-07 | | Release date: | 2001-11-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III.
Cell(Cambridge,Mass.), 106, 2001
|
|
1YEY
 
 | | Crystal Structure of L-fuconate Dehydratase from Xanthomonas campestris pv. campestris str. ATCC 33913 | | Descriptor: | L-fuconate dehydratase, MAGNESIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-28 | | Release date: | 2005-01-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal Structure of L-fuconate Dehydratase from Xanthomonas campestris pv. campestris str. ATCC 33913
To be Published
|
|
4E09
 
 | | Structure of ParF-AMPPCP, I422 form | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Plasmid partitioning protein ParF, SULFATE ION | | Authors: | Schumacher, M.A, Ye, Q, Barge, M.R, Barilla, D, Hayes, F. | | Deposit date: | 2012-03-02 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural Mechanism of ATP-induced Polymerization of the Partition Factor ParF: IMPLICATIONS FOR DNA SEGREGATION.
J.Biol.Chem., 287, 2012
|
|
4EK1
 
 | | Crystal Structure of Electron-Spin Labeled Cytochrome P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Lee, Y.-T, Goodin, D.B. | | Deposit date: | 2012-04-08 | | Release date: | 2012-07-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Double electron-electron resonance shows cytochrome P450cam undergoes a conformational change in solution upon binding substrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
