4RS7
 
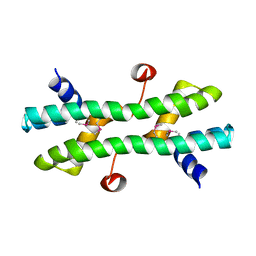 | |
4RS8
 
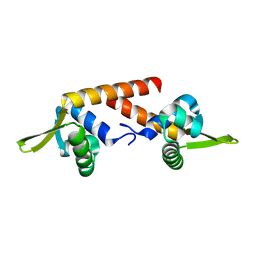 | |
1P94
 
 | | NMR Structure of ParG symmetric dimer | | Descriptor: | plasmid partition protein ParG | | Authors: | Golovanov, A.P, Barilla, D, Golovanova, M, Hayes, F, Lian, L.Y. | | Deposit date: | 2003-05-09 | | Release date: | 2004-01-13 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | ParG, a protein required for active partition of bacterial plasmids, has a dimeric ribbon-helix-helix structure.
Mol.Microbiol., 50, 2003
|
|
4E03
 
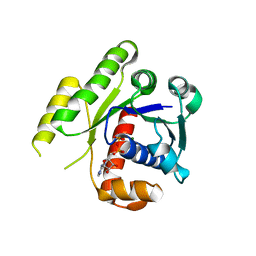 | | Structure of ParF-ADP form 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Plasmid partitioning protein ParF | | Authors: | Schumacher, M.A, Ye, Q, Barge, M.R, Barilla, D, Hayes, F. | | Deposit date: | 2012-03-02 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Mechanism of ATP-induced Polymerization of the Partition Factor ParF: IMPLICATIONS FOR DNA SEGREGATION.
J.Biol.Chem., 287, 2012
|
|
4DZZ
 
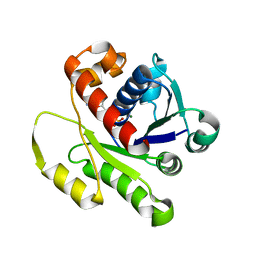 | | Structure of ParF-ADP, crystal form 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Plasmid partitioning protein ParF | | Authors: | Schumacher, M.A, Ye, Q, Barge, M.R, Barilla, D, Hayes, F. | | Deposit date: | 2012-03-01 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Mechanism of ATP-induced Polymerization of the Partition Factor ParF: IMPLICATIONS FOR DNA SEGREGATION.
J.Biol.Chem., 287, 2012
|
|
4E07
 
 | | ParF-AMPPCP-C2221 form | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Plasmid partitioning protein ParF | | Authors: | Schumacher, M.A, Ye, Q, Barge, M.R, Barilla, D, Hayes, F. | | Deposit date: | 2012-03-02 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Mechanism of ATP-induced Polymerization of the Partition Factor ParF: IMPLICATIONS FOR DNA SEGREGATION.
J.Biol.Chem., 287, 2012
|
|
4E09
 
 | | Structure of ParF-AMPPCP, I422 form | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Plasmid partitioning protein ParF, SULFATE ION | | Authors: | Schumacher, M.A, Ye, Q, Barge, M.R, Barilla, D, Hayes, F. | | Deposit date: | 2012-03-02 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural Mechanism of ATP-induced Polymerization of the Partition Factor ParF: IMPLICATIONS FOR DNA SEGREGATION.
J.Biol.Chem., 287, 2012
|
|
