5KBT
 
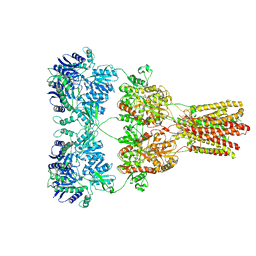 | | Cryo-EM structure of GluA2-1xSTZ complex at 6.4 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy.
Science, 353, 2016
|
|
1W0K
 
 | | ADP inhibited bovine F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE ALPHA CHAIN HEART ISOFORM, MITOCHONDRIAL PRECURSOR, ... | | Authors: | Kagawa, R, Montgomery, M.G, Braig, K, Walker, J.E, Leslie, A.G.W. | | Deposit date: | 2004-06-08 | | Release date: | 2004-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Structure of Bovine F1-ATPase Inhibited by Adp and Beryllium Fluoride
Embo J., 23, 2004
|
|
1W0J
 
 | | Beryllium fluoride inhibited bovine F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE ALPHA CHAIN HEART ISOFORM, MITOCHONDRIAL PRECURSOR, ... | | Authors: | Kagawa, R, Montgomery, M.G, Braig, K, Walker, J.E, Leslie, A.G.W. | | Deposit date: | 2004-06-08 | | Release date: | 2004-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of Bovine F1-ATPase Inhibited by Adp and Beryllium Fluoride
Embo J., 23, 2004
|
|
4PBE
 
 | | Phosphotriesterase Variant Rev6 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-revR6, ... | | Authors: | Campbell, E, Kaltenbach, M, Tokuriki, N, Jackson, C.J. | | Deposit date: | 2014-04-12 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat. Chem. Biol., 12, 2016
|
|
2ORQ
 
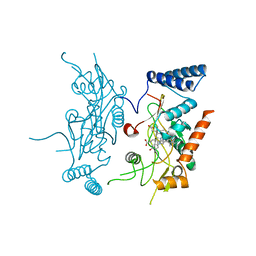 | | Murine Inducible Nitric Oxide Synthase Oxygenase Domain (DELTA 114) 4-(imidazol-1-yl)phenol and piperonylamine Complex | | Descriptor: | 1-(1,3-BENZODIOXOL-5-YL)METHANAMINE, 4-(1H-IMIDAZOL-1-YL)PHENOL, Nitric oxide synthase, ... | | Authors: | Adler, M, Whitlow, M. | | Deposit date: | 2007-02-04 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, Synthesis, and Activity of 2-Imidazol-1-ylpyrimidine Derived Inducible Nitric Oxide Synthase Dimerization Inhibitors
J.Med.Chem., 50, 2007
|
|
2OO4
 
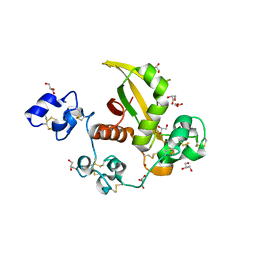 | | Structure of LNR-HD (Negative Regulatory Region) from human Notch 2 | | Descriptor: | CALCIUM ION, GLYCEROL, Neurogenic locus notch homolog protein 2, ... | | Authors: | Gordon, W.R, Vardar-Ulu, D, Histen, G, Sanchez-Irizarry, C, Aster, J.C, Blacklow, S.C. | | Deposit date: | 2007-01-25 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural basis for autoinhibition of Notch
Nat.Struct.Mol.Biol., 14, 2007
|
|
2ORP
 
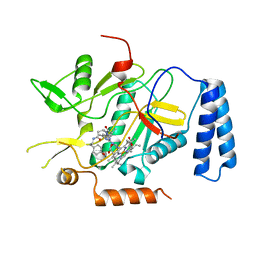 | | Murine inducible nitric oxide synthase oxygenase domain (delta 114) 2-[4-(2-Imidazol-1-yl-6-methyl-pyrimidin-4-yl)-1-isobutyryl-piperazin-2-yl]-N-[2-(4-methoxy-phenyl)-ethyl]-acetamide complex | | Descriptor: | 2-{(2R)-4-[2-(1H-IMIDAZOL-1-YL)-6-METHYLPYRIMIDIN-4-YL]-1-ISOBUTYRYLPIPERAZIN-2-YL}-N-[2-(4-METHOXYPHENYL)ETHYL]ACETAMIDE, PROTOPORPHYRIN IX CONTAINING FE, nitric oxide synthase, ... | | Authors: | Adler, M, Whitlow, M. | | Deposit date: | 2007-02-04 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Design, Synthesis, and Activity of 2-Imidazol-1-ylpyrimidine Derived Inducible Nitric Oxide Synthase Dimerization Inhibitors
J.Med.Chem., 50, 2007
|
|
2HXY
 
 | | Crystal structure of human apo-eIF4AIII | | Descriptor: | Probable ATP-dependent RNA helicase DDX48 | | Authors: | Johansen, J.S, Andersen, G.R. | | Deposit date: | 2006-08-04 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA.
Science, 313, 2006
|
|
437D
 
 | | CRYSTAL STRUCTURE OF AN RNA PSEUDOKNOT FROM BEET WESTERN YELLOW VIRUS INVOLVED IN RIBOSOMAL FRAMESHIFTING | | Descriptor: | MAGNESIUM ION, RNA PSEUDOKNOT, SODIUM ION | | Authors: | Su, L, Chen, L, Egli, M, Berger, J.M, Rich, A. | | Deposit date: | 1998-11-24 | | Release date: | 1998-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Minor groove RNA triplex in the crystal structure of a ribosomal frameshifting viral pseudoknot.
Nat.Struct.Biol., 6, 1999
|
|
1Y8O
 
 | | Crystal structure of the PDK3-L2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DIHYDROLIPOIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, ... | | Authors: | Kato, M, Chuang, J.L, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2004-12-13 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of pyruvate dehydrogenase kinase 3 bound to lipoyl domain 2 of human pyruvate dehydrogenase complex.
Embo J., 24, 2005
|
|
2CDV
 
 | | REFINED STRUCTURE OF CYTOCHROME C3 AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Higuchi, Y, Kusunoki, M, Matsuura, Y, Yasuoka, N, Kakudo, M. | | Deposit date: | 1983-11-15 | | Release date: | 1984-02-02 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined structure of cytochrome c3 at 1.8 A resolution
J.Mol.Biol., 172, 1984
|
|
1J9S
 
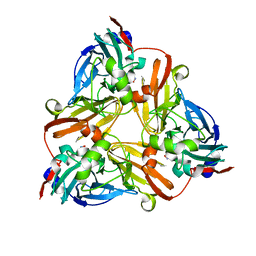 | | Crystal structure of nitrite soaked oxidized H255N AFNIR | | Descriptor: | COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, NITRITE ION | | Authors: | Boulanger, M.J, Murphy, M.E. | | Deposit date: | 2001-05-28 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alternate substrate binding modes to two mutant (D98N and H255N) forms of nitrite reductase from Alcaligenes faecalis S-6: structural model of a transient catalytic intermediate
Biochemistry, 40, 2001
|
|
1VYM
 
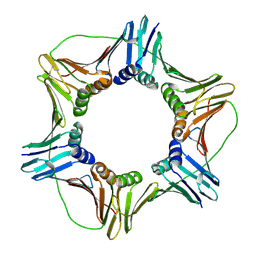 | | NATIVE HUMAN PCNA | | Descriptor: | PROLIFERATING CELL NUCLEAR ANTIGEN | | Authors: | Kontopidis, G, Wu, S, Zheleva, D, Taylor, P, Mcinnes, C, Lane, D, Fischer, P, Walkinshaw, M. | | Deposit date: | 2004-05-03 | | Release date: | 2005-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Studies of Human Proliferating Cell Nuclear Antigen Complexes Provide a Rationale for Cyclin Association and Inhibitor Design
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1VYJ
 
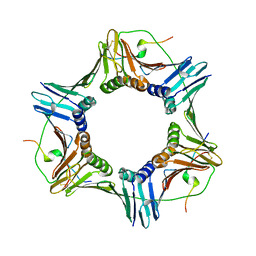 | | Structural and biochemical studies of human PCNA complexes provide the basis for association with CDK/cyclin and rationale for inhibitor design | | Descriptor: | PROLIFERATING CELL NUCLEAR ANTIGEN, SMALL PEPTIDE SAVLQKKITDYFHPKK | | Authors: | Kontopidis, G, Wu, S, Zheleva, D, Taylor, P, Mcinnes, C, Lane, D, Fischer, P, Walkinshaw, M.D. | | Deposit date: | 2004-04-30 | | Release date: | 2005-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Biochemical Studies of Human Proliferating Cell Nuclear Antigen Complexes Provide a Rationale for Cyclin Association and Inhibitor Design
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
4RUZ
 
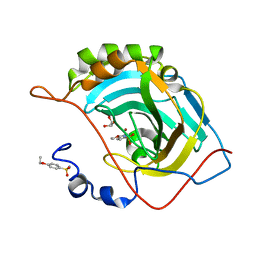 | |
3UTV
 
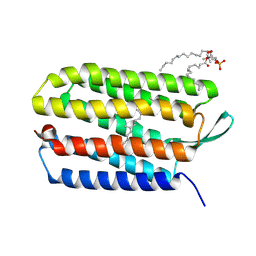 | | Crystal structure of bacteriorhodopsin mutant Y57F | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Bacteriorhodopsin, RETINAL | | Authors: | Cao, Z, Bowie, J.U. | | Deposit date: | 2011-11-26 | | Release date: | 2012-05-09 | | Last modified: | 2014-07-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Shifting hydrogen bonds may produce flexible transmembrane helices.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2CY3
 
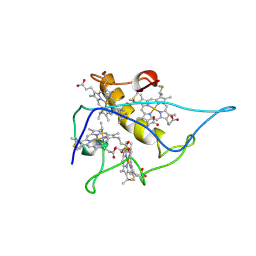 | |
1J9T
 
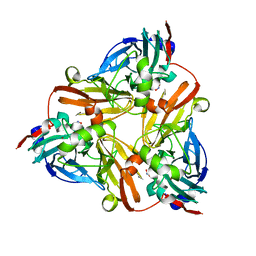 | | Crystal structure of nitrite soaked reduced H255N AFNIR | | Descriptor: | COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, NITRITE ION | | Authors: | Boulanger, M.J, Murphy, M.E. | | Deposit date: | 2001-05-28 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Alternate substrate binding modes to two mutant (D98N and H255N) forms of nitrite reductase from Alcaligenes faecalis S-6: structural model of a transient catalytic intermediate
Biochemistry, 40, 2001
|
|
423D
 
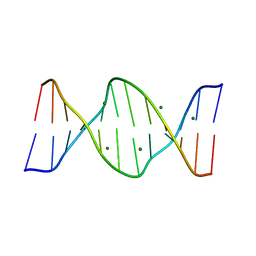 | | 5'-D(*AP*CP*CP*GP*AP*CP*GP*TP*CP*GP*GP*T)-3' | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*AP*CP*GP*TP*CP*GP*GP*T)-3'), MAGNESIUM ION | | Authors: | Rozenberg, H, Rabinovich, D, Frolow, F, Hegde, R.S, Shakked, Z. | | Deposit date: | 1998-09-14 | | Release date: | 1999-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural code for DNA recognition revealed in crystal structures of papillomavirus E2-DNA targets.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
455D
 
 | |
2P1L
 
 | |
1FKC
 
 | | HUMAN PRION PROTEIN (MUTANT E200K) FRAGMENT 90-231 | | Descriptor: | PRION PROTEIN | | Authors: | Zhang, Y, Swietnicki, W, Zagorski, M.G, Surewicz, W.K, Soennichsen, F.D. | | Deposit date: | 2000-08-09 | | Release date: | 2000-09-21 | | Last modified: | 2018-03-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the E200K variant of human prion protein. Implications for the mechanism of pathogenesis in familial prion diseases.
J.Biol.Chem., 275, 2000
|
|
4DYX
 
 | |
4Q3O
 
 | | Crystal structure of MGS-MT1, an alpha/beta hydrolase enzyme from a Lake Matapan deep-sea metagenome library | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
7PXV
 
 | | LsAA9_A chemically reduced with ascorbic acid (high X-ray dose) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
