6V64
 
 | | Crystal structure of human thrombin bound to ppack with tryptophans replaced by 5-F-tryptophan | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, Thrombin heavy chain, ... | | Authors: | Ruben, E.A, Chen, Z, Di Cera, E. | | Deposit date: | 2019-12-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | 19F NMR reveals the conformational properties of free thrombin and its zymogen precursor prethrombin-2.
J.Biol.Chem., 295, 2020
|
|
6CFD
 
 | | ADEP4 bound to E. faecium ClpP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit, N-[(6aS,12S,15aS,17R,21R,23aS)-17,21-dimethyl-6,11,15,20,23-pentaoxooctadecahydro-2H,6H,11H,15H-pyrido[2,1-i]dipyrrolo[2,1-c:2',1'-l][1,4,7,10,13]oxatetraazacyclohexadecin-12-yl]-3,5-difluoro-Nalpha-[(2E)-hept-2-enoyl]-L-phenylalaninamide | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2018-02-14 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | In VivoandIn VitroEffects of a ClpP-Activating Antibiotic against Vancomycin-Resistant Enterococci.
Antimicrob. Agents Chemother., 62, 2018
|
|
1HJK
 
 | | ALKALINE PHOSPHATASE MUTANT H331Q | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Murphy, J.E, Stec, B, Ma, L, Kantrowitz, E.R. | | Deposit date: | 1997-05-30 | | Release date: | 1997-10-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Trapping and visualization of a covalent enzyme-phosphate intermediate.
Nat.Struct.Biol., 4, 1997
|
|
5MG0
 
 | | Structure of PAS-GAF fragment of Deinococcus phytochrome by serial femtosecond crystallography | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome, ... | | Authors: | Burgie, E.S, Fuller, F.D, Gul, S, Miller, M.D, Young, I.D, Brewster, A.S, Clinger, J, Aller, P, Braeuer, P, Hutchison, C, Alonso-Mori, R, Kern, J, Yachandra, V.K, Yano, J, Sauter, N.K, Phillips Jr, G.N, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2016-11-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers.
Nat. Methods, 14, 2017
|
|
8CO3
 
 | |
6P7Q
 
 | | Structure of E. coli MS115-1 NucC, 5'-pApA bound form | | Descriptor: | CHLORIDE ION, E. coli MS115-1 NucC, RNA (5'-R(P*AP*A)-3') | | Authors: | Ye, Q, Lau, R.K, Berg, K.R, Corbett, K.D. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
1ACW
 
 | | SOLUTION NMR STRUCTURE OF P01, A NATURAL SCORPION PEPTIDE STRUCTURALLY ANALOGOUS TO SCORPION TOXINS SPECIFIC FOR APAMIN-SENSITIVE POTASSIUM CHANNEL, 25 STRUCTURES | | Descriptor: | NATURAL SCORPION PEPTIDE P01 | | Authors: | Blanc, E, Fremont, V, Sizun, P, Meunier, S, Van Rietschoten, J, Thevand, A, Bernassau, J.M, Darbon, H. | | Deposit date: | 1997-02-10 | | Release date: | 1997-04-01 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of P01, a natural scorpion peptide structurally analogous to scorpion toxins specific for apamin-sensitive potassium channel.
Proteins, 24, 1996
|
|
3CAB
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera soaked at pH 7.0 | | Descriptor: | GLYCEROL, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-02-19 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
7ZHG
 
 | | High-resolution cryo-EM structure of Pyrococcus abyssi 30S ribosomal subunit bound to mRNA and initiator tRNA anticodon stem-loop | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Kazan, R, Bourgeois, G, Mechulam, Y, Coureux, P.D, Schmitt, E. | | Deposit date: | 2022-04-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.25 Å) | | Cite: | Role of aIF5B in archaeal translation initiation.
Nucleic Acids Res., 50, 2022
|
|
3CDN
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera soaked at pH 4.0 | | Descriptor: | CHLORIDE ION, GLYCEROL, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-02-27 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
7Q1D
 
 | | Acetyltrasferase(3) type IIIa in complex with 3-N-methyl-nemycin B | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aminoglycoside N(3)-acetyltransferase III, CHLORIDE ION, ... | | Authors: | Pontillo, N, Guskov, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | 3-N-alkylation in aminoglycoside antibiotic neomycin B overcomes bacterial resistance mediated by acetyltransferase (3) IIIa
To Be Published
|
|
3CZ1
 
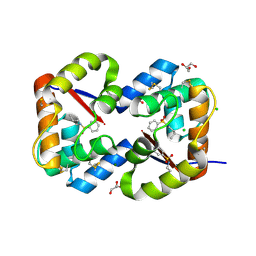 | | Dimeric crystal structure of a pheromone binding protein from Apis mellifera in complex with the n-butyl benzene sulfonamide at pH 7.0 | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-04-27 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
7Q4K
 
 | | Erythromycin-stalled Escherichia coli 70S ribosome with streptococcal MsrDL nascent chain | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fostier, C.R, Ousalem, F, Soufari, H, Leroy, E.C, Ngo, S, Innis, A, Hashem, Y, Boel, G. | | Deposit date: | 2021-10-31 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Regulation of the macrolide resistance ABC-F translation factor MsrD.
Nat Commun, 14, 2023
|
|
6PYD
 
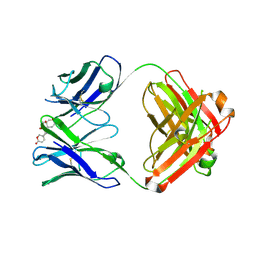 | | Structure of 3E9 antibody Fab bound to marinobufagenin | | Descriptor: | (3beta,5beta,14alpha,15beta)-3,5-dihydroxy-14,15-epoxybufa-20,22-dienolide, 3E9 anti-marinobufagenin antibody Fab heavy chain, recloned with human IgG4 C region, ... | | Authors: | Franklin, M.C, Macdonald, L.E, McWhirter, J, Murphy, A.J. | | Deposit date: | 2019-07-29 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kappa-on-Heavy (KoH) bodies are a distinct class of fully-human antibody-like therapeutic agents with antigen-binding properties.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7T82
 
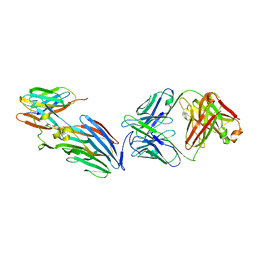 | | Crystal Structure of LEUKOCIDIN E/CENTYRIN S26/FAB B438 | | Descriptor: | Antibody Fab Heavy Chain, Antibody Fab Light Chain, Centyrin S26, ... | | Authors: | Luo, J, Malia, T.J, Buckley, P.T. | | Deposit date: | 2021-12-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Multivalent human antibody-centyrin fusion protein to prevent and treat Staphylococcus aureus infections.
Cell Host Microbe, 31, 2023
|
|
4QUN
 
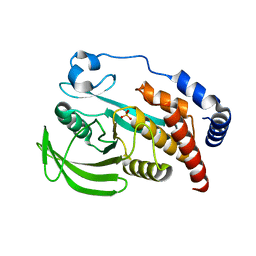 | |
7QFP
 
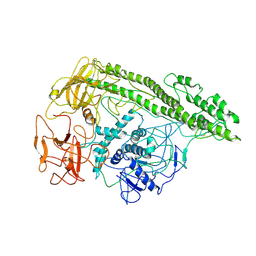 | | Cryo-EM structure of Botulinum neurotoxin serotype E | | Descriptor: | Botulinum neurotoxin | | Authors: | Kosenina, S, Martinez-Carranza, M, Davies, J.R, Masuyer, G, Stenmark, P. | | Deposit date: | 2021-12-06 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Analysis of Botulinum Neurotoxins Type B and E by Cryo-EM.
Toxins, 14, 2021
|
|
7QGM
 
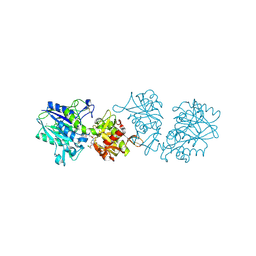 | |
7QGL
 
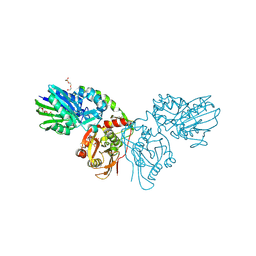 | |
7QGO
 
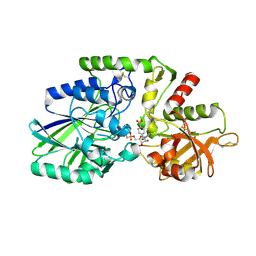 | | Human CD73 (ecto 5'-nucleotidase) in complex with MRS4602 (a 3-methyl-CMPCP derivative, compound 21 in paper) in the closed state (crystal form III) | | Descriptor: | 5'-nucleotidase, ZINC ION, [[(2~{R},3~{S},4~{R},5~{R})-5-[(4~{E})-4-[(4-methoxycarbonylphenyl)methoxyimino]-3-methyl-2-oxidanylidene-pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]methylphosphonic acid | | Authors: | Strater, N. | | Deposit date: | 2021-12-09 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structure-Activity Relationship of 3-Methylcytidine-5'-alpha , beta-methylenediphosphates as CD73 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7QGA
 
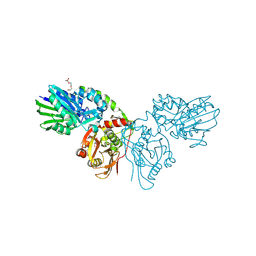 | |
6GDR
 
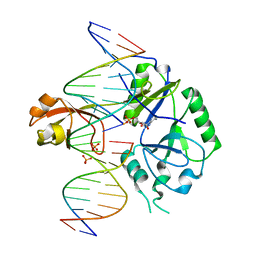 | | DNA binding with a minimal scaffold: Structure-function analysis of Lig E DNA ligases | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA, DNA (5'-D(*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*T)-3'), ... | | Authors: | Williamson, A, Grigic, M, Leiros, H.K.S. | | Deposit date: | 2018-04-24 | | Release date: | 2018-07-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | DNA binding with a minimal scaffold: structure-function analysis of Lig E DNA ligases.
Nucleic Acids Res., 46, 2018
|
|
4RG8
 
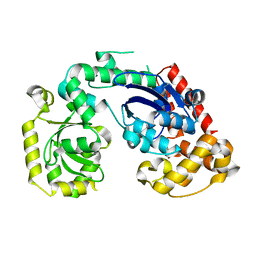 | | Structural and biochemical studies of a moderately thermophilic Exonuclease I from Methylocaldum szegediense | | Descriptor: | Exonuclease I, MAGNESIUM ION | | Authors: | Fei, L, Tian, S, Moysey, R, Misca, M, Barker, J.J, Smith, M.A, McEwan, P.A, Pilka, E.S, Crawley, L, Evans, T, Sun, D. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural and Biochemical Studies of a Moderately Thermophilic Exonuclease I from Methylocaldum szegediense.
Plos One, 10, 2015
|
|
1B9G
 
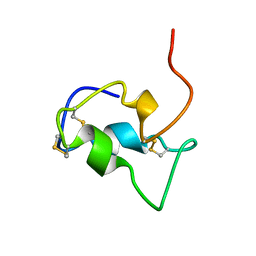 | | INSULIN-LIKE-GROWTH-FACTOR-1 | | Descriptor: | PROTEIN (GROWTH FACTOR IGF-1) | | Authors: | De Wolf, E, Gill, R, Geddes, S, Pitts, J, Wollmer, A, Grotzinger, J. | | Deposit date: | 1999-02-11 | | Release date: | 1999-02-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a mini IGF-1.
Protein Sci., 5, 1996
|
|
5KQZ
 
 | | Protease E35D-CaP2 | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, Protease | | Authors: | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
