6T6M
 
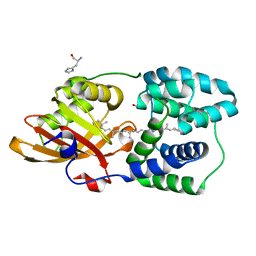 | | Y201W mutant of the orange carotenoid protein from Synechocystis at pH 5.5 | | Descriptor: | GLYCEROL, HISTIDINE, Orange carotenoid-binding protein, ... | | Authors: | Sluchanko, N.N, Gushchin, I, Botnarevskiy, V.S, Slonimskiy, Y.B, Remeeva, A, Kovalev, K, Stepanov, A.V, Gordeliy, V, Maksimov, E.G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Role of hydrogen bond alternation and charge transfer states in photoactivation of the Orange Carotenoid Protein.
Commun Biol, 4, 2021
|
|
6S08
 
 | | Crystal Structure of Properdin (TSR domains N1 & 456) | | Descriptor: | Properdin, SODIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | van den Bos, R.M, Pearce, N.M, Gros, P. | | Deposit date: | 2019-06-14 | | Release date: | 2019-09-04 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Insights Into Enhanced Complement Activation by Structures of Properdin and Its Complex With the C-Terminal Domain of C3b.
Front Immunol, 10, 2019
|
|
3MDD
 
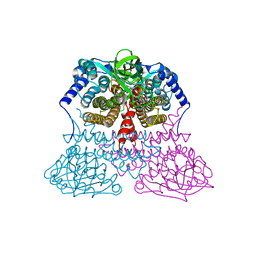 | |
3MDE
 
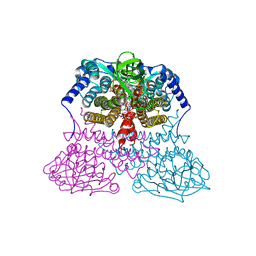 | |
4AMV
 
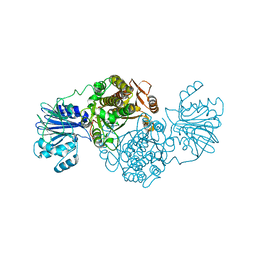 | |
6BH8
 
 | | Crystal structure of ZMPSTE24 in complex with phosphoramidon | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CAAX prenyl protease 1 homolog, N-ALPHA-L-RHAMNOPYRANOSYLOXY(HYDROXYPHOSPHINYL)-L-LEUCYL-L-TRYPTOPHAN, ... | | Authors: | Goblirsch, B.R, Arachea, B.T, Wiener, M.C. | | Deposit date: | 2017-10-30 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Phosphoramidon inhibits the integral membrane protein zinc metalloprotease ZMPSTE24.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
8UTX
 
 | | Solution structure of a 12-mer peptide bearing a bicyclic Asx motif mimic (BAMM) as a synthetic N-cap | | Descriptor: | 1,3,5-tris(bromomethyl)benzene, TRP-CYS-ASP-ALA-ALA-CYS-CYS-ALA-ALA-ALA-LYS-ALA-NH2 peptide | | Authors: | Mi, T.X, Burgess, K. | | Deposit date: | 2023-10-31 | | Release date: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Bioinformatics leading to conveniently accessible, helix enforcing, bicyclic ASX motif mimics (BAMMs).
Nat Commun, 15, 2024
|
|
4AOW
 
 | | Crystal structure of the human Rack1 protein at a resolution of 2.45 angstrom | | Descriptor: | GLYCEROL, GUANINE NUCLEOTIDE-BINDING PROTEIN SUBUNIT BETA-2-LIKE 1 | | Authors: | Ruiz Carrillo, D, Chandrasekaran, R, Nilsson, M, Cornvick, T, Liew, C.W, Tan, S.M, Lescar, J. | | Deposit date: | 2012-03-30 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of Human Rack1 Protein at a Resolution of 2.45 A.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2M0P
 
 | | Solution structure of the tenth complement type repeat of human megalin | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor-related protein 2 | | Authors: | Dagil, R, Kragelund, B. | | Deposit date: | 2012-11-01 | | Release date: | 2013-01-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Gentamicin binds to the megalin receptor as a competitive inhibitor using the common ligand binding motif of complement type repeats: insight from the nmr structure of the 10th complement type repeat domain alone and in complex with gentamicin.
J.Biol.Chem., 288, 2013
|
|
2M4X
 
 | |
2EVQ
 
 | |
2MJ9
 
 | | Designed Exendin-4 analogues | | Descriptor: | Exendin-4 | | Authors: | Rovo, P, Farkas, V, Straner, P, Szabo, M, Jermendy, A, Hegyi, O, Toth, G.K, Perczel, A. | | Deposit date: | 2013-12-30 | | Release date: | 2014-06-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Rational design of alpha-helix-stabilized exendin-4 analogues.
Biochemistry, 53, 2014
|
|
2M50
 
 | | Analysis of the structural and molecular basis of voltage-sensitive sodium channel inhibition by the spider toxin, Huwentoxin-IV (-TRTX-Hh2a). | | Descriptor: | Mu-theraphotoxin-Hh2a | | Authors: | Gibbs, A, Minassian, N, Flinspach, M, Wickenden, A. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | Analysis of the Structural and Molecular Basis of Voltage-sensitive Sodium Channel Inhibition by the Spider Toxin Huwentoxin-IV ( mu-TRTX-Hh2a).
J.Biol.Chem., 288, 2013
|
|
3NYZ
 
 | | Crystal Structure of Kemp Elimination Catalyst 1A53-2 | | Descriptor: | Indole-3-glycerol phosphate synthase, SULFATE ION | | Authors: | Lee, T.M, Privett, H.K, Kaiser, J.T, Mayo, S.L. | | Deposit date: | 2010-07-15 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.514 Å) | | Cite: | Iterative approach to computational enzyme design.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3NZ1
 
 | | Crystal Structure of Kemp Elimination Catalyst 1A53-2 Complexed with Transition State Analog 5-Nitro Benzotriazole | | Descriptor: | 5-nitro-1H-benzotriazole, Indole-3-glycerol phosphate synthase, L(+)-TARTARIC ACID, ... | | Authors: | Lee, T.M, Privett, H.K, Kaiser, J.T, Mayo, S.L. | | Deposit date: | 2010-07-15 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Iterative approach to computational enzyme design.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1A22
 
 | | HUMAN GROWTH HORMONE BOUND TO SINGLE RECEPTOR | | Descriptor: | GROWTH HORMONE, GROWTH HORMONE RECEPTOR | | Authors: | De Vos, A.M, Ultsch, M. | | Deposit date: | 1998-01-15 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional analysis of the 1:1 growth hormone:receptor complex reveals the molecular basis for receptor affinity.
J.Mol.Biol., 277, 1998
|
|
6YNN
 
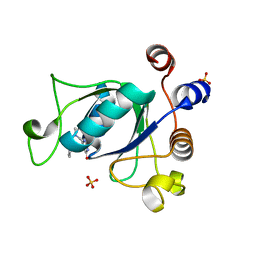 | | Crystal structure of YTHDC1 with compound DHU_DC1_135 | | Descriptor: | 6-[[(2-chloranyl-6-fluoranyl-phenyl)methyl-methyl-amino]methyl]-1~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6YNO
 
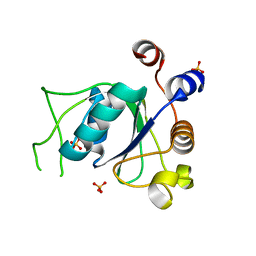 | | Crystal structure of YTHDC1 with compound DHU_DC1_139 | | Descriptor: | 6-[[methyl-[(1-phenylpyrazol-3-yl)methyl]amino]methyl]-1~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
2V5V
 
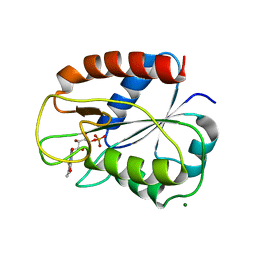 | | W57E Flavodoxin from Anabaena | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, MAGNESIUM ION | | Authors: | Herguedas, B, Martinez-Julvez, M, Perez-Dorado, I, Goni, G, Medina, M, Hermoso, J.A. | | Deposit date: | 2007-07-10 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Tuning of the Fmn Binding and Oxido-Reduction Properties by Neighboring Side Chains in Anabaena Flavodoxin.
Arch.Biochem.Biophys., 467, 2007
|
|
6ZCN
 
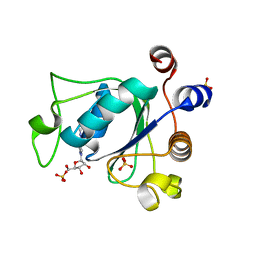 | | Crystal structure of YTHDC1 with m6A | | Descriptor: | N6-METHYLADENOSINE-5'-MONOPHOSPHATE, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6ZCM
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_180 | | Descriptor: | 6-[[cyclopropyl-[(7-methoxy-1,3-benzodioxol-5-yl)methyl]amino]methyl]-1~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
7L18
 
 | | Crystal structure of a tandem deletion mutant of rat NADPH-cytochrome P450 reductase | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hubbard, P.A, Xia, C, Shen, A.L, Kim, J.J.K. | | Deposit date: | 2020-12-14 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.542 Å) | | Cite: | Structural and kinetic investigations of the carboxy terminus of NADPH-cytochrome P450 oxidoreductase.
Arch.Biochem.Biophys., 701, 2021
|
|
2WA8
 
 | | Structural basis of N-end rule substrate recognition in Escherichia coli by the ClpAP adaptor protein ClpS - The Phe peptide structure | | Descriptor: | ATP-DEPENDENT CLP PROTEASE ADAPTER PROTEIN CLPS, N-END RULE PEPTIDE | | Authors: | Schuenemann, V.J, Kralik, S.M, Albrecht, R, Spall, S.K, Truscott, K.N, Dougan, D.A, Zeth, K. | | Deposit date: | 2009-02-03 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis of N-End Rule Substrate Recognition in Escherichia Coli by the Clpap Adaptor Protein Clps.
Embo Rep., 10, 2009
|
|
4NUY
 
 | | Crystal structure of EndoS, an endo-beta-N-acetyl-glucosaminidase from Streptococcus pyogenes | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase F2 | | Authors: | Trastoy, B, Guenther, S, Snyder, G.A, Sundberg, E.J. | | Deposit date: | 2013-12-04 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Crystal structure of Streptococcus pyogenes EndoS, an immunomodulatory endoglycosidase specific for human IgG antibodies.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3CSM
 
 | | STRUCTURE OF YEAST CHORISMATE MUTASE WITH BOUND TRP AND AN ENDOOXABICYCLIC INHIBITOR | | Descriptor: | 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, CHORISMATE MUTASE, TRYPTOPHAN | | Authors: | Straeter, N, Schnappauf, G, Braus, G, Lipscomb, W.N. | | Deposit date: | 1997-07-10 | | Release date: | 1998-01-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanisms of catalysis and allosteric regulation of yeast chorismate mutase from crystal structures.
Structure, 5, 1997
|
|
