6NUB
 
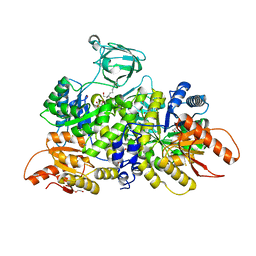 | | Pyruvate Kinase M2 Mutant - S437Y in Complex with L-serine | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Srivastava, D, Nandi, S, Dey, M. | | Deposit date: | 2019-01-31 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanistic and Structural Insights into Cysteine-Mediated Inhibition of Pyruvate Kinase Muscle Isoform 2.
Biochemistry, 58, 2019
|
|
1F29
 
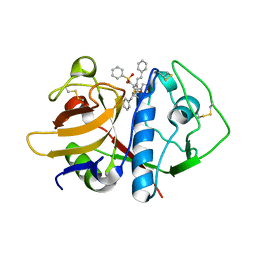 | | CRYSTAL STRUCTURE ANALYSIS OF CRUZAIN BOUND TO A VINYL SULFONE DERIVED INHIBITOR (I) | | Descriptor: | 3-[[N-[MORPHOLIN-N-YL]-CARBONYL]-PHENYLALANINYL-AMINO]-5- PHENYL-PENTANE-1-SULFONYLBENZENE, CRUZAIN | | Authors: | Brinen, L.S, Hansell, E, Roush, W.R, McKerrow, J.H, Fletterick, R.J. | | Deposit date: | 2000-05-23 | | Release date: | 2000-07-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A target within the target: probing cruzain's P1' site to define structural determinants for the Chagas' disease protease.
Structure Fold.Des., 8, 2000
|
|
1AG0
 
 | |
1F2A
 
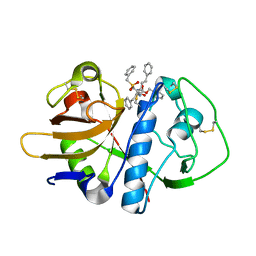 | | CRYSTAL STRUCTURE ANALYSIS OF CRUZAIN BOUND TO A VINYL SULFONE DERIVED INHIBITOR (II) | | Descriptor: | 3-[N-[BENZYLOXYCARBONYL]-PHENYLALANINYL-AMINO]-5-PHENYL-PENTANE-1-SULFONYLMETHYLBENZENE, CRUZAIN | | Authors: | Brinen, L.S, Hansell, E, Roush, W.R, McKerrow, J.H, Fletterick, R.J. | | Deposit date: | 2000-05-23 | | Release date: | 2000-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A target within the target: probing cruzain's P1' site to define structural determinants for the Chagas' disease protease.
Structure Fold.Des., 8, 2000
|
|
1F2B
 
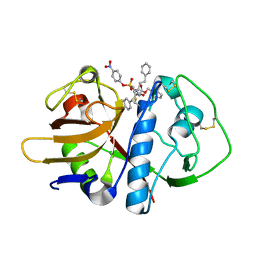 | | CRYSTAL STRUCTURE ANALYSIS OF CRUZAIN BOUND TO VINYL SULFONE DERIVED INHIBITOR (III) | | Descriptor: | 3-[N-[BENZYLOXYCARBONYL]-PHENYLALANINYL-AMINO]-5-PHENYL-PENTANE-1-SULFONIC ACID 4-NITRO-PHENYL ESTER, CRUZAIN | | Authors: | Brinen, L.S, Hansell, E, Roush, W.R, McKerrow, J.H, Fletterick, R.J. | | Deposit date: | 2000-05-23 | | Release date: | 2000-07-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A target within the target: probing cruzain's P1' site to define structural determinants for the Chagas' disease protease.
Structure Fold.Des., 8, 2000
|
|
1F2C
 
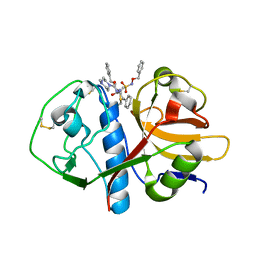 | | CRYSTAL STRUCTURE ANALYSIS OF CRYZAIN BOUND TO VINYL SULFONE DERIVED INHIBITOR (IV) | | Descriptor: | 3-[[N-[4-METHYL-PIPERAZINYL]CARBONYL]-PHENYLALANINYL-AMINO]-5-PHENYL-PENTANE-1-SULFONIC ACID BENZYLOXY-AMIDE, CRUZAIN | | Authors: | Brinen, L.S, Hansell, E, Roush, W.R, McKerrow, J.H, Fletterick, R.J. | | Deposit date: | 2000-05-23 | | Release date: | 2000-07-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A target within the target: probing cruzain's P1' site to define structural determinants for the Chagas' disease protease.
Structure Fold.Des., 8, 2000
|
|
2P86
 
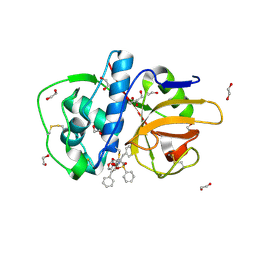 | | The high resolution crystal structure of rhodesain, the major cathepsin L protease from T. brucei rhodesiense, bound to inhibitor K11002 | | Descriptor: | 1,2-ETHANEDIOL, 3-[[N-[MORPHOLIN-N-YL]-CARBONYL]-PHENYLALANINYL-AMINO]-5- PHENYL-PENTANE-1-SULFONYLBENZENE, Cysteine protease | | Authors: | Brinen, L.S, Marion, R. | | Deposit date: | 2007-03-21 | | Release date: | 2008-04-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | THE high resolution structure of rhodesain, the major cathepsin L protease from trypanosoma brucei rhodesiense, illustrates the basis for differences in inhibition profiles from other papain family cysteine proteases
To be Published
|
|
8GSK
 
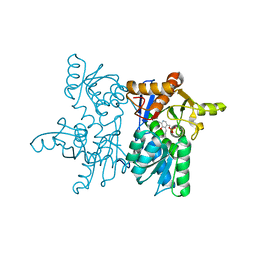 | |
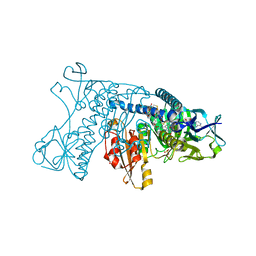
4GR1
 
 | |
7YOL
 
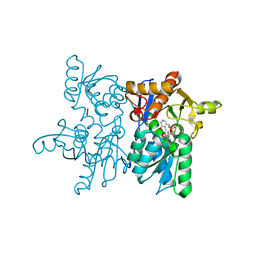 | | Crystal structure of tetra mutant (D67E, A68P, L98I, A301S) of O-acetylserine sulfhydrylase from Haemophilus influenzae in complex with high-affinity inhibitory peptide of serine acetyltransferase from Haemophilus influenzae at 2.4 A | | Descriptor: | Cysteine synthase, Peptide from Serine acetyltransferase | | Authors: | Saini, N, Kumar, N, Rahisuddin, R, Singh, A.K. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of tetra mutant (D67E, A68P, L98I, A301S) of O-acetylserine sulfhydrylase from Haemophilus influenzae in complex with high-affinity inhibitory peptide from serine acetyltransferase of Haemophilus influenzae at 2.4 A
To Be Published
|
|
8EG6
 
 | | huCaspase-6 in complex with inhibitor 2a | | Descriptor: | (3R)-1-(ethanesulfonyl)-N-[4-(trifluoromethoxy)phenyl]piperidine-3-carboxamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Zhao, Y, Fan, P, Liu, J, Wang, Y, Van Horn, K, Wang, D, Medina-Cleghorn, D, Lee, P, Bryant, C, Altobelli, C, Jaishankar, P, Ng, R.A, Ambrose, A.J, Tang, Y, Arkin, M.R, Renslo, A.R. | | Deposit date: | 2022-09-11 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Engaging a Non-catalytic Cysteine Residue Drives Potent and Selective Inhibition of Caspase-6.
J.Am.Chem.Soc., 145, 2023
|
|
8EG5
 
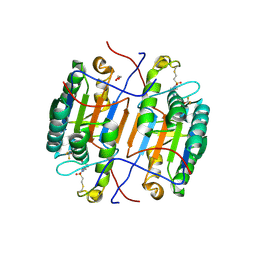 | | huCaspase-6 in complex with inhibitor 3a | | Descriptor: | (3R,5S)-1-(ethanesulfonyl)-5-phenyl-N-[4-(trifluoromethoxy)phenyl]piperidine-3-carboxamide (bound form), 1,2-ETHANEDIOL, Caspase-6 subunit p11, ... | | Authors: | Zhao, Y, Fan, P, Liu, J, Wang, Y, Van Horn, K, Wang, D, Medina-Cleghorn, D, Lee, P, Bryant, C, Altobelli, C, Jaishankar, P, Ng, R.A, Ambrose, A.J, Tang, Y, Arkin, M.R, Renslo, A.R. | | Deposit date: | 2022-09-11 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Engaging a Non-catalytic Cysteine Residue Drives Potent and Selective Inhibition of Caspase-6.
J.Am.Chem.Soc., 145, 2023
|
|
6UKD
 
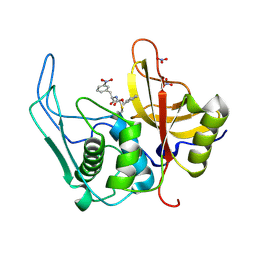 | |
4TVV
 
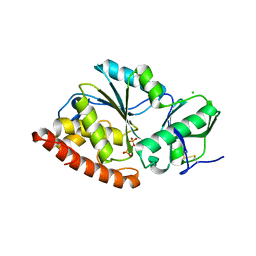 | | Crystal structure of LppA from Legionella pneumophila | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Weber, S, Stirnimann, C, Wieser, M, Meier, R, Engelhardt, S, Li, X, Capitani, G, Kammerer, R, Hilbi, H. | | Deposit date: | 2014-06-28 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Type IV Translocated Legionella Cysteine Phytase Counteracts Intracellular Growth Restriction by Phytate.
J.Biol.Chem., 289, 2014
|
|
4ORE
 
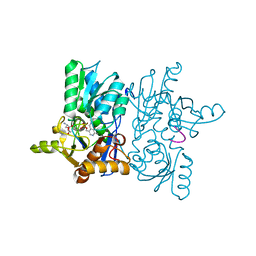 | |
2POA
 
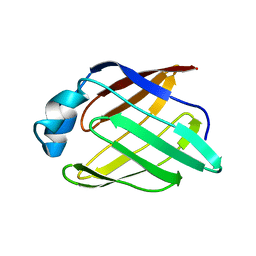 | | Schistosoma mansoni Sm14 Fatty Acid-Binding Protein: improvement of protein stability by substitution of the single Cys62 residue | | Descriptor: | 14 kDa fatty acid-binding protein | | Authors: | Ramos, C.R.R, Oyama Jr, S, Sforca, M.L, Pertinhez, T.A, Ho, P.L, Spisni, A. | | Deposit date: | 2007-04-26 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Stability improvement of the fatty acid binding protein Sm14 from S. mansoni by Cys replacement: Structural and functional characterization of a vaccine candidate.
Biochim.Biophys.Acta, 1794, 2009
|
|
8JCR
 
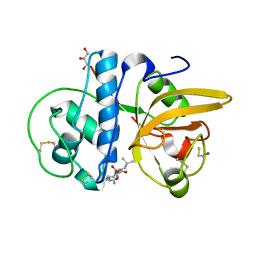 | | Crystal structure of Calotropain FI from Calotropis gigantea (pH 6.0) | | Descriptor: | GLYCEROL, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, Procerain, ... | | Authors: | Kumar, A, Jamdar, S.N, Srivastava, G, Makde, R.D. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-22 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Papain-Like Cysteine Protease, Calotropain FI, Purified from the Latex of Calotropis gigantea.
J.Agric.Food Chem., 73, 2025
|
|
8JCQ
 
 | | Crystal structure of calotropain FI from Calotropis gigantea | | Descriptor: | N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, Procerain | | Authors: | Kumar, A, Jamdar, S.N, Srivastava, G, Makde, R.D. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-22 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of Papain-Like Cysteine Protease, Calotropain FI, Purified from the Latex of Calotropis gigantea.
J.Agric.Food Chem., 73, 2025
|
|
7YOF
 
 | |
7YOE
 
 | |
7YOD
 
 | |
7YOG
 
 | |
7YOK
 
 | | Crystal Structure of Tetra mutant (D67E, A68P, L98I, A301S) tetra mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae at 2.8 A | | Descriptor: | Cysteine synthase | | Authors: | Saini, N, Kumar, N, Rahisuddin, R, Singh, A.K. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Crystal Structure of Tetra mutant (D67E, A68P, L98I, A301S) tetra mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae at 2.8 A
To Be Published
|
|
7YOI
 
 | |
7YOM
 
 | | Crystal structure of tetra mutant (D67E,A68P,L98I,A301S) of O-acetylserine sulfhydrylase from Salmonella typhimurium in complex with high-affinity inhibitory peptide from serine acetyltransferase of Salmonella typhimurium at 2.8 A | | Descriptor: | Cysteine synthase, peptide from serine acetyltransferase | | Authors: | Saini, N, Kumar, N, Rahisuddin, R, Singh, A.K. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of I88L single mutant of O-acetylserine sulfhydrylase from Haemophilus influenzae in complex with high-affinity inhibitory peptide from serine acetyltransferase of Salmonella typhimurium at 2.14 A
To Be Published
|
|
