8FC2
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, hygromycin A, and azithromycin at 2.50A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Syroegin, E.A, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the mechanism of overcoming Erm-mediated resistance by macrolides acting together with hygromycin-A.
Nat Commun, 14, 2023
|
|
8CLH
 
 | | Drug cocktail (Colchicine, Epothilone A, Peloruside, Ansamitocin P3, Vinblastine) bound to tubulin (T2R-TTL) complex | | Descriptor: | (1S,2S,3S,5S,6S,16Z,18Z,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl 2-methylpropanoate, (2ALPHA,2'BETA,3BETA,4ALPHA,5BETA)-VINCALEUKOBLASTINE, CALCIUM ION, ... | | Authors: | Wranik, M, Bertrand, Q, Kepa, M.W, Weinert, T, Steinmetz, M, Standfuss, J. | | Deposit date: | 2023-02-16 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A multi-reservoir extruder for time-resolved serial protein crystallography and compound screening at X-ray free-electron lasers.
Nat Commun, 14, 2023
|
|
8FC5
 
 | | Crystal structure of the A2058-N6-dimethylated Thermus thermophilus 70S ribosome in complex with protein Y, hygromycin A, and azithromycin at 2.65A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Syroegin, E.A, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-26 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into the mechanism of overcoming Erm-mediated resistance by macrolides acting together with hygromycin-A.
Nat Commun, 14, 2023
|
|
8C0Z
 
 | | CryoEM structure of a tungsten-containing aldehyde oxidoreductase from Aromatoleum aromaticum | | Descriptor: | Aldehyde:ferredoxin oxidoreductase,tungsten-containing, BENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Winiarska, A, Ramirez-Amador, F, Hege, D, Gemmecker, Y, Prinz, S, Hochberg, G, Heider, J, Szaleniec, M, Schuller, J.M. | | Deposit date: | 2022-12-19 | | Release date: | 2023-05-31 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | A bacterial tungsten-containing aldehyde oxidoreductase forms an enzymatic decorated protein nanowire.
Sci Adv, 9, 2023
|
|
8FC6
 
 | | Crystal structure of the A2058-N6-dimethylated Thermus thermophilus 70S ribosome in complex with protein Y, hygromycin A, and telithromycin at 2.35A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Syroegin, E.A, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-26 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into the mechanism of overcoming Erm-mediated resistance by macrolides acting together with hygromycin-A.
Nat Commun, 14, 2023
|
|
4Z8A
 
 | | SH3-III of Drosophila Rim-binding protein bound to a Cacophony derived peptide | | Descriptor: | RIM-binding protein, isoform F, Voltage-dependent calcium channel type A subunit alpha-1 | | Authors: | Driller, J.H, Holton, N, Siebert, M, Boehme, A.M, Wahl, M.C, Sigrist, S.J, Loll, B. | | Deposit date: | 2015-04-08 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | A high affinity RIM-binding protein/Aplip1 interaction prevents the formation of ectopic axonal active zones.
Elife, 4, 2015
|
|
6ZVJ
 
 | | Structure of a human ABCE1-bound 43S pre-initiation complex - State II | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Kratzat, H, Mackens-Kiani, T, Ameismeier, A, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes.
Embo J., 40, 2021
|
|
5KDJ
 
 | | ZmpB metallopeptidase from Clostridium perfringens | | Descriptor: | F5/8 type C domain protein, GLYCEROL, SODIUM ION, ... | | Authors: | Noach, I, Ficko-Blean, E, Stuart, C, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6FQM
 
 | | 3.06A COMPLEX OF S.AUREUS GYRASE with imidazopyrazinone T1 AND DNA | | Descriptor: | 7-[(3~{S})-3-azanylpyrrolidin-1-yl]-5-cyclopropyl-8-fluoranyl-imidazo[1,2-a]quinoxalin-4-one, DNA (5'-D(*GP*AP*GP*AP*GP*TP*AP*T*GP*GP*CP*CP*AP*TP*AP*CP*TP*CP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Germe, T, Basque, E, Maxwell, A. | | Deposit date: | 2018-02-14 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | A new class of antibacterials, the imidazopyrazinones, reveal structural transitions involved in DNA gyrase poisoning and mechanisms of resistance.
Nucleic Acids Res., 46, 2018
|
|
3KF7
 
 | | Crystal Structure of Human p38alpha Complexed With a Triazolopyrimidine compound | | Descriptor: | 3-{6-[2-(2,4-difluorophenyl)ethyl][1,2,4]triazolo[4,3-a]pyridin-3-yl}-4-methylbenzamide, Mitogen-activated protein kinase 14 | | Authors: | Shieh, H.-S, Williams, J.M, Stegeman, R.A, Xing, L, Jerome, K.D. | | Deposit date: | 2009-10-27 | | Release date: | 2009-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Continued exploration of the triazolopyridine scaffold as a platform for p38 MAP kinase inhibition.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6PEG
 
 | | MIF with a allosteric inhibitor | | Descriptor: | 4-amino-5-hydroxy-6-[(E)-(3-{[3-(2-methylpropanoyl)pyrazolo[1,5-a]pyridin-2-yl]methyl}phenyl)diazenyl]naphthalene-1,3-disulfonic acid, ACETATE ION, GLYCEROL, ... | | Authors: | Asojo, O.A. | | Deposit date: | 2019-06-20 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of Macrophage Migration Inhibitory Factor by a Chimera of Two Allosteric Binders.
Acs Med.Chem.Lett., 11, 2020
|
|
7QNE
 
 | | Cryo-EM structure of human full-length synaptic alpha1beta3gamma2 GABA(A)R in complex with Ro15-4513 and megabody Mb38 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Sente, A, Desai, R, Naydenova, K, Malinauskas, T, Jounaidi, Y, Miehling, J, Zhou, X, Masiulis, S, Hardwick, S.W, Chirgadze, D.Y, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Differential assembly diversifies GABA A receptor structures and signalling.
Nature, 604, 2022
|
|
9C9L
 
 | | Cryo-EM structure of the C1q A, B-crt, C peptide assembly narrow region | | Descriptor: | Peptide from Complement C1q subcomponent subunit A, Peptide from Complement C1q subcomponent subunit B, Peptide from Complement C1q subcomponent subunit C | | Authors: | Kreutzberger, M.A, Yu, L.T, Egelman, E.H, Hartgerink, J.D. | | Deposit date: | 2024-06-14 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A Collagen Triple Helix without the Superhelical Twist.
Acs Cent.Sci., 11, 2025
|
|
6TU5
 
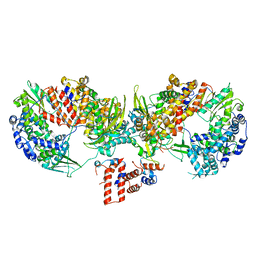 | | Influenza A/H7N9 polymerase core (apo) | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Cusack, S, Pflug, A. | | Deposit date: | 2020-01-02 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.325 Å) | | Cite: | A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase.
Cell, 181, 2020
|
|
8EQM
 
 | | Structure of a dimeric photosystem II complex acclimated to far-red light | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Gisriel, C.J, Shen, G, Flesher, D.A, Kurashov, V, Golbeck, J.H, Brudvig, G.W, Amin, M, Bryant, D.A. | | Deposit date: | 2022-10-08 | | Release date: | 2022-12-28 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of a dimeric photosystem II complex from a cyanobacterium acclimated to far-red light.
J.Biol.Chem., 299, 2022
|
|
8Z82
 
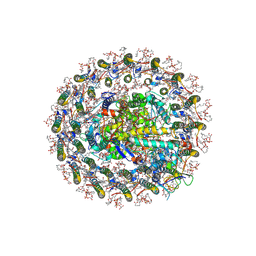 | | Photosynthetic LH1-RC-HiPIP complex from the purple bacterium Halorhodospira halophila | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, Antenna complex, ... | | Authors: | Tani, K, Kanno, R, Nagashima, K.V.P, Hiwatashi, N, Kawakami, M, Nakata, K, Nagashima, S, Inoue, K, Takaichi, S, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2024-04-21 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | A Native LH1-RC-HiPIP Supercomplex from an Extremophilic Phototroph.
Commun Biol, 8, 2025
|
|
8Z83
 
 | | Photosynthetic LH1-RC complex from the purple bacterium Halorhodospira halophila | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, Antenna complex, ... | | Authors: | Tani, K, Kanno, R, Nagashima, K.V.P, Hiwatashi, N, Kawakami, M, Nakata, K, Nagashima, S, Inoue, K, Takaichi, S, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2024-04-21 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A Native LH1-RC-HiPIP Supercomplex from an Extremophilic Phototroph.
Commun Biol, 8, 2025
|
|
5LMK
 
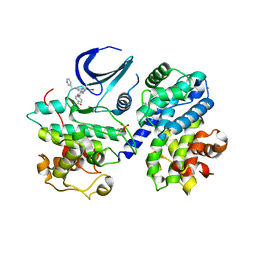 | | Structure of phopsho-CDK2-cyclin A in complex with an ATP-competitive inhibitor | | Descriptor: | 4-[4-[3-bromanyl-7-(pyridin-3-ylmethylamino)pyrazolo[1,5-a]pyrimidin-5-yl]phenyl]benzamide, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Echalier, A. | | Deposit date: | 2016-08-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Explicit treatment of active-site waters enhances quantum mechanical/implicit solvent scoring: Inhibition of CDK2 by new pyrazolo[1,5-a]pyrimidines.
Eur J Med Chem, 126, 2016
|
|
8Z81
 
 | | Photosynthetic LH2-LH1 complex from the purple bacterium Halorhodospira halophila | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Antenna complex, alpha/beta subunit, ... | | Authors: | Tani, K, Nagashima, K.V.P, Kanno, R, Hiwatashi, N, Kawakami, M, Nakata, K, Nagashima, S, Inoue, K, Takaichi, S, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2024-04-21 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | A distinct double-ring LH1-LH2 photocomplex from an extremophilic phototroph.
Nat Commun, 16, 2025
|
|
9DIP
 
 | | Crystal structure of H5 hemagglutinin from the influenza virus A/Texas/37/2024 (H5N1) with LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Lin, T.H, Zhu, Y, Wilson, I.A. | | Deposit date: | 2024-09-05 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A single mutation in bovine influenza H5N1 hemagglutinin switches specificity to human receptors.
Science, 386, 2024
|
|
6QWH
 
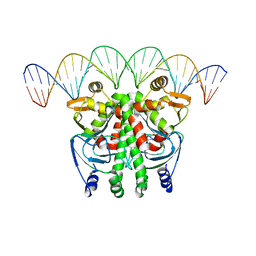 | | The Transcriptional Regulator PrfA-L140H mutant from Listeria Monocytogenes in complex with a 30-bp operator PrfA-box motif | | Descriptor: | DNA (30-MER), GLYCEROL, Listeriolysin positive regulatory factor A | | Authors: | Hall, M, Grundstrom, C, Hansen, S, Brannstrom, K, Johansson, J, Sauer-Eriksson, A.E. | | Deposit date: | 2019-03-05 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Novel Growth-Based Selection Strategy Identifies New Constitutively Active Variants of the Major Virulence Regulator PrfA in Listeria monocytogenes.
J.Bacteriol., 202, 2020
|
|
6QWF
 
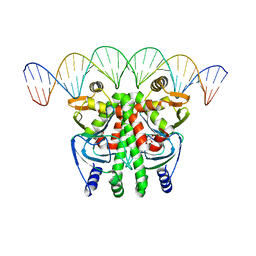 | | The Transcriptional Regulator PrfA-A94V mutant from Listeria Monocytogenes in complex with a 30-bp operator PrfA-box motif | | Descriptor: | DNA (30-MER), GLYCEROL, Listeriolysin positive regulatory factor A | | Authors: | Hall, M, Grundstrom, C, Hansen, S, Brannstrom, K, Johansson, J, Sauer-Eriksson, A.E. | | Deposit date: | 2019-03-05 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Novel Growth-Based Selection Strategy Identifies New Constitutively Active Variants of the Major Virulence Regulator PrfA in Listeria monocytogenes.
J.Bacteriol., 202, 2020
|
|
6QWM
 
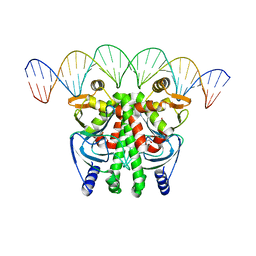 | | The Transcriptional Regulator PrfA-A218G mutant from Listeria Monocytogenes in complex with a 30-bp operator PrfA-box motif | | Descriptor: | DNA (30-MER), GLYCEROL, Listeriolysin positive regulatory factor A | | Authors: | Hall, M, Grundstrom, C, Hansen, S, Brannstrom, K, Johansson, J, Sauer-Eriksson, A.E. | | Deposit date: | 2019-03-05 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Novel Growth-Based Selection Strategy Identifies New Constitutively Active Variants of the Major Virulence Regulator PrfA in Listeria monocytogenes.
J.Bacteriol., 202, 2020
|
|
7ZY6
 
 | |
8A9N
 
 | | Structure of DpA polyamine acetyltransferase in complex with 1,3-DAP | | Descriptor: | 1,3-DIAMINOPROPANE, Acetyltransferase, COENZYME A, ... | | Authors: | Garcia-Pino, A, Jurenas, D. | | Deposit date: | 2022-06-28 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | A polyamine acetyltransferase regulates the motility and biofilm formation of Acinetobacter baumannii.
Nat Commun, 14, 2023
|
|
