5ARN
 
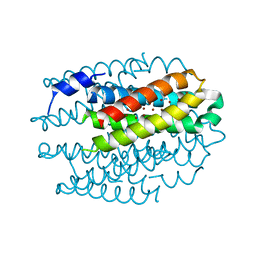 | | Cu(I)-CSP3 (COPPER STORAGE PROTEIN 3) FROM METHYLOSINUS | | Descriptor: | COPPER (I) ION, CSP3 | | Authors: | Vita, N, Landolfi, G, Basle, A, Platsaki, S, Waldron, K, Dennison, C. | | Deposit date: | 2015-09-25 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel Cytosolic Copper Storage Proteins
To be Published
|
|
6U7M
 
 | |
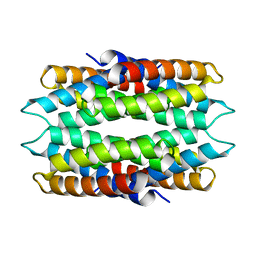
6WKT
 
 | |
7TH0
 
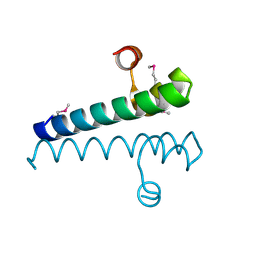 | | Escherichia coli RpnA-S | | Descriptor: | Recombination-promoting nuclease RpnA | | Authors: | Zhong, A, Hickman, A.B, Storz, G, Dyda, F. | | Deposit date: | 2022-01-10 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Toxic antiphage defense proteins inhibited by intragenic antitoxin proteins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5L8B
 
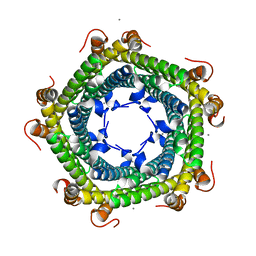 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant E62A | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2016-06-07 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments.
Elife, 5, 2016
|
|
5L89
 
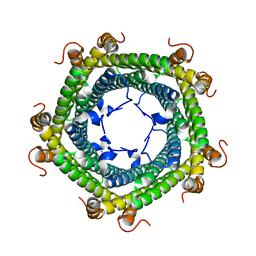 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant E32A | | Descriptor: | CALCIUM ION, Rru_A0973 | | Authors: | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2016-06-07 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments.
Elife, 5, 2016
|
|
4FYP
 
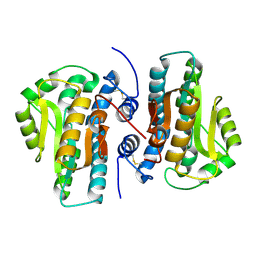 | | Crystal Structure of Plant Vegetative Storage Protein | | Descriptor: | MAGNESIUM ION, Vegetative storage protein 1 | | Authors: | Chen, Y, Wei, J, Wang, M, Gong, W, Zhang, M. | | Deposit date: | 2012-07-05 | | Release date: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of Arabidopsis VSP1 reveals the plant class C-like phosphatase structure of the DDDD superfamily of phosphohydrolases
Plos One, 7, 2012
|
|
7KG1
 
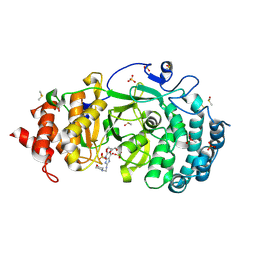 | | Structure of human PARG complexed with PARG-002 | | Descriptor: | 1,3-dimethyl-8-{[2-(morpholin-4-yl)ethyl]amino}-3,7-dihydro-1H-purine-2,6-dione, CACODYLATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Brosey, C.A, Balapiti-Modarage, L.P.F, Warden, L.S, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-15 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
7KG7
 
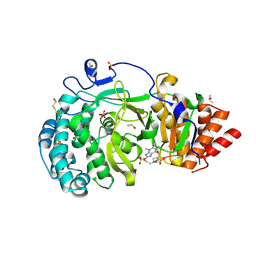 | | Structure of human PARG complexed with PARG-292 | | Descriptor: | 8-{[2-(1,1-dioxo-1lambda~6~,4-thiazinan-4-yl)ethyl]sulfanyl}-1,3-dimethyl-3,7-dihydro-1H-purine-2,6-dione, DIMETHYL SULFOXIDE, Poly(ADP-ribose) glycohydrolase, ... | | Authors: | Brosey, C.A, Balapiti-Modarage, L.P.F, Warden, L.S, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-16 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
7KG6
 
 | | Structure of human PARG complexed with PARG-322 | | Descriptor: | 1-{2-[(1,3-dimethyl-2,6-dioxo-2,3,6,7-tetrahydro-1H-purin-8-yl)sulfanyl]ethyl}piperidine-4-carboxylic acid, Poly(ADP-ribose) glycohydrolase | | Authors: | Brosey, C.A, Balapiti-Modarage, L.P.F, Warden, L.S, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-16 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
7KG8
 
 | | Structure of human PARG complexed with PARG-061 | | Descriptor: | 1,3-dimethyl-8-{[2-(morpholin-4-yl)-2-oxoethyl]sulfanyl}-6-sulfanylidene-1,3,6,7-tetrahydro-2H-purin-2-one, CACODYLATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Brosey, C.A, Balapiti-Modarage, L.P.F, Warden, L.S, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-16 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
5L8G
 
 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant H65A | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2016-06-07 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.974 Å) | | Cite: | Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments.
Elife, 5, 2016
|
|
6ZBI
 
 | | Ternary complex of Calmodulin bound to 2 molecules of NHE1 | | Descriptor: | CALCIUM ION, Calmodulin-1, Sodium/hydrogen exchanger 1 | | Authors: | Prestel, A, Kragelund, B.B, Pedersen, E.S, Pedersen, S.F, Sjoegaard-Frich, L.M. | | Deposit date: | 2020-06-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dynamic Na + /H + exchanger 1 (NHE1) - calmodulin complexes of varying stoichiometry and structure regulate Ca 2+ -dependent NHE1 activation.
Elife, 10, 2021
|
|
5MAM
 
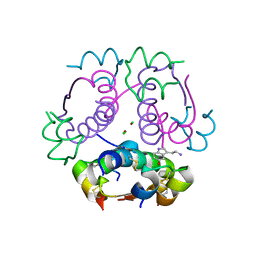 | | Human insulin in complex with serotonin | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Brzozowski, A.M, Turkenburg, J.P, Jiracek, J, Zakova, L. | | Deposit date: | 2016-11-03 | | Release date: | 2017-04-05 | | Last modified: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Computational and structural evidence for neurotransmitter-mediated modulation of the oligomeric states of human insulin in storage granules.
J. Biol. Chem., 292, 2017
|
|
5MT3
 
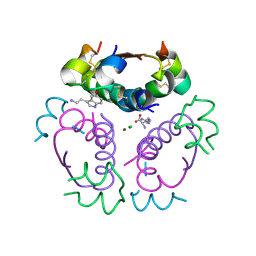 | | Human insulin in complex with serotonin and arginine | | Descriptor: | ARGININE, CHLORIDE ION, Insulin, ... | | Authors: | Brzozowski, A.M, Turkenburg, J.P, Jiracek, J, Zakova, L. | | Deposit date: | 2017-01-06 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Computational and structural evidence for neurotransmitter-mediated modulation of the oligomeric states of human insulin in storage granules.
J. Biol. Chem., 292, 2017
|
|
5MT9
 
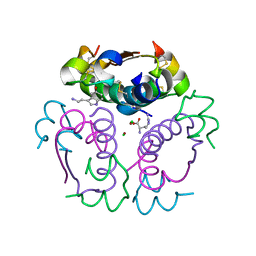 | | Human insulin in complex with serotonin and arginine | | Descriptor: | ARGININE, CHLORIDE ION, Insulin, ... | | Authors: | Brzozowski, A.M, Turkenburg, J.P, Jiracek, J, Zakova, L. | | Deposit date: | 2017-01-07 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Computational and structural evidence for neurotransmitter-mediated modulation of the oligomeric states of human insulin in storage granules.
J. Biol. Chem., 292, 2017
|
|
7AC1
 
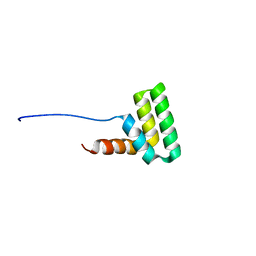 | | Solution structure of the TAF4-RST domain | | Descriptor: | Transcription initiation factor TFIID subunit 4 | | Authors: | Staby, L, Davidsen, R, Bugge, K, Skriver, K, Kragelund, B.B. | | Deposit date: | 2020-09-09 | | Release date: | 2021-10-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | alpha alpha-hub coregulator structure and flexibility determine transcription factor binding and selection in regulatory interactomes.
J.Biol.Chem., 298, 2022
|
|
4KZ7
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 16 ((1R,4S)-4,7,7-trimethyl-3-oxo-2-oxabicyclo[2.2.1]heptane-1-carboxylic acid) | | Descriptor: | (1R,4S)-4,7,7-trimethyl-3-oxo-2-oxabicyclo[2.2.1]heptane-1-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZ6
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 13 ((2R,6R)-6-methyl-1-(3-sulfanylpropanoyl)piperidine-2-carboxylic acid) | | Descriptor: | (2R,6R)-6-methyl-1-(3-sulfanylpropanoyl)piperidine-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZ9
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 41 ((4R,4aS,8aS)-4-phenyldecahydroquinolin-4-ol) | | Descriptor: | (4R,4aS,8aS)-4-phenyldecahydroquinolin-4-ol, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZA
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 48 (3-(cyclopropylsulfamoyl)thiophene-2-carboxylic acid) | | Descriptor: | 3-(cyclopropylsulfamoyl)thiophene-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZ3
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 44 (5-chloro-3-sulfamoylthiophene-2-carboxylic acid) | | Descriptor: | 5-chloro-3-sulfamoylthiophene-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZB
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 50 (N-(methylsulfonyl)-N-phenyl-alanine) | | Descriptor: | Beta-lactamase, N-(methylsulfonyl)-N-phenyl-D-alanine, N-(methylsulfonyl)-N-phenyl-L-alanine | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZ4
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 60 (2-[(propylsulfonyl)amino]benzoic acid) | | Descriptor: | 2-[(propylsulfonyl)amino]benzoic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZ8
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 20 (1,3-diethyl-2-thioxodihydropyrimidine-4,6(1H,5H)-dione) | | Descriptor: | 1,3-diethyl-2-thioxodihydropyrimidine-4,6(1H,5H)-dione, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
