5E17
 
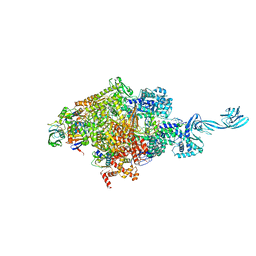 | |
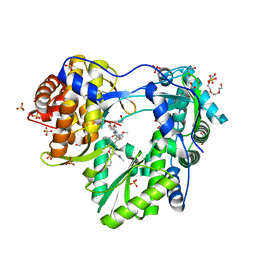
5QJ1
 
 | |
5NRG
 
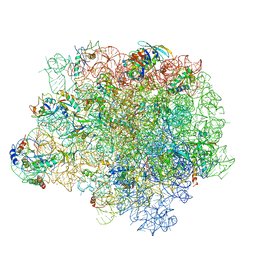 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus in complex with RB02 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S ribosomal RNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Yonath, A, Matzov, D, Eyal, Z, Ben Hamou, R, Zimmerman, E, Rozenberg, H, Bashan, A, Fridman, M. | | Deposit date: | 2017-04-23 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.442 Å) | | Cite: | Structural insights of lincosamides targeting the ribosome of Staphylococcus aureus.
Nucleic Acids Res., 45, 2017
|
|
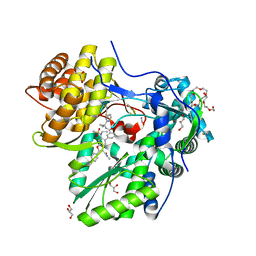
5QJ0
 
 | |
6IFV
 
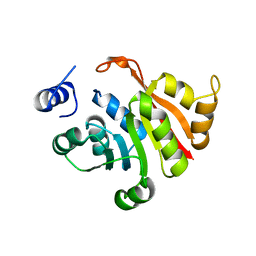 | | C-terminal truncated KsgA from Bacillus subtilis 168 | | Descriptor: | Ribosomal RNA small subunit methyltransferase A | | Authors: | Bhujbalrao, R, Anand, R. | | Deposit date: | 2018-09-21 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Deciphering Determinants in Ribosomal Methyltransferases That Confer Antimicrobial Resistance.
J. Am. Chem. Soc., 141, 2019
|
|
8H3V
 
 | | Cryo-EM structure of the full transcription activation complex NtcA-NtcB-TAC | | Descriptor: | DNA (125-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Han, S.J, Jiang, Y.L, You, L.L, Shen, L.Q, Wu, X.X, Yang, F, Kong, W.W, Chen, Z.P, Zhang, Y, Zhou, C.Z. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | DNA looping mediates cooperative transcription activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H40
 
 | | Cryo-EM structure of the transcription activation complex NtcA-TAC | | Descriptor: | DNA (125-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Han, S.J, Jiang, Y.L, You, L.L, Shen, L.Q, Wu, X.X, Yang, F, Kong, W.W, Chen, Z.P, Zhang, Y, Zhou, C.Z. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | DNA looping mediates cooperative transcription activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6IFX
 
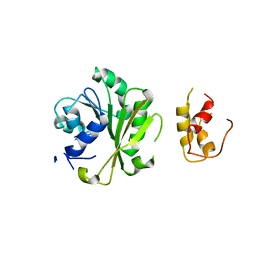 | |
7UO4
 
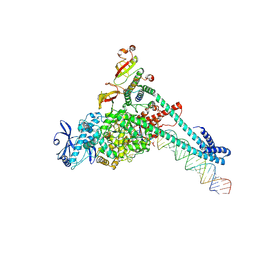 | | SARS-CoV-2 replication-transcription complex bound to Remdesivir triphosphate, in a pre-catalytic state | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UOB
 
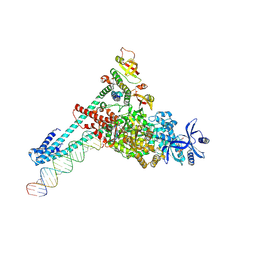 | | SARS-CoV-2 replication-transcription complex bound to GTP, in a pre-catalytic state | | Descriptor: | 3'-DEOXYURIDINE-5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UO9
 
 | | SARS-CoV-2 replication-transcription complex bound to UTP, in a pre-catalytic state | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UO7
 
 | | SARS-CoV-2 replication-transcription complex bound to ATP, in a pre-catalytic state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UOE
 
 | | SARS-CoV-2 replication-transcription complex bound to CTP, in a pre-catalytic state | | Descriptor: | 3'-DEOXYURIDINE-5'-MONOPHOSPHATE, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
1HNW
 
 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH TETRACYCLINE | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Brodersen, D.E, Clemons Jr, W.M, Carter, A.P, Morgan-Warren, R, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-08 | | Release date: | 2001-02-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit.
Cell(Cambridge,Mass.), 103, 2000
|
|
6R9G
 
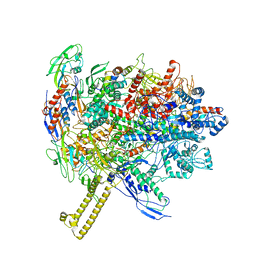 | |
1HNZ
 
 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH HYGROMYCIN B | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Brodersen, D.E, Clemons Jr, W.M, Carter, A.P, Morgan-Warren, R, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-08 | | Release date: | 2001-02-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit.
Cell(Cambridge,Mass.), 103, 2000
|
|
5E18
 
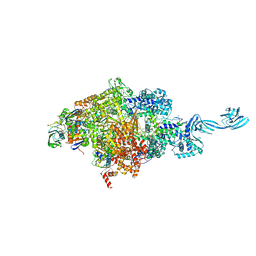 | |
6PB6
 
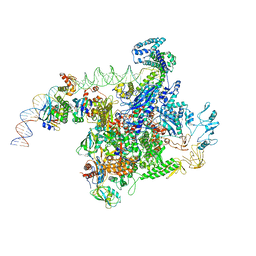 | |
8H2H
 
 | | Cryo-EM structure of a Group II Intron Complexed with its Reverse Transcriptase | | Descriptor: | Group II intron-encoded protein LtrA, LtrB, RNA (5'-R(P*CP*AP*CP*AP*UP*CP*CP*AP*UP*AP*AP*C)-3') | | Authors: | Liu, N, Dong, X.L, Qu, G.S, Wang, J, Wang, H.W, Belfort, M. | | Deposit date: | 2022-10-06 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Functionalized graphene grids with various charges for single-particle cryo-EM.
Nat Commun, 13, 2022
|
|
8EBU
 
 | | XPC release from Core7-XPA-DNA (Cy5) | | Descriptor: | DNA repair protein complementing XP-A cells, DNA repair protein complementing XP-C cells, DNA1, ... | | Authors: | Kim, J, Yang, W. | | Deposit date: | 2022-08-31 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Lesion recognition by XPC, TFIIH and XPA in DNA excision repair.
Nature, 617, 2023
|
|
8EBY
 
 | | XPC release from Core7-XPA-DNA (AP) | | Descriptor: | DNA, DNA repair protein complementing XP-A cells, DNA repair protein complementing XP-C cells, ... | | Authors: | Kim, J, Yang, W. | | Deposit date: | 2022-08-31 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Lesion recognition by XPC, TFIIH and XPA in DNA excision repair.
Nature, 617, 2023
|
|
8FVR
 
 | | CryoEM structure of E.coli transcription elongation complex | | Descriptor: | DNA (53-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Duan, W, Serganov, A. | | Deposit date: | 2023-01-19 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Control of transcription elongation and DNA repair by alarmone ppGpp.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FVW
 
 | |
7AIH
 
 | | The Large subunit of the Kinetoplastid mitochondrial ribosome | | Descriptor: | 50S ribosomal protein L13-like protein, C2H2-type domain-containing protein, L51_S25_CI-B8 domain-containing protein, ... | | Authors: | Soufari, H, Waltz, F, Parrot, C, Bochler, A, Hashem, Y. | | Deposit date: | 2020-09-27 | | Release date: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the mature kinetoplastids mitoribosome and insights into its large subunit biogenesis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PB5
 
 | |
