6OGQ
 
 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P30) in complex with GRL-003 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(4-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
6OGS
 
 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P30) in complex with GRL-001 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
6OYD
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-004 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-1-(4-fluorophenyl)-3-hydroxy-4-[(2-methylpropyl)({2-[(propan-2-yl)amino]-1,3-benzoxazol-6-yl}sulfonyl)amino]butan-2-yl}carbamate, Protease | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-05-14 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
6OGR
 
 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P30) in complex with GRL-142 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, 1,2-ETHANEDIOL, Protease | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Novel HIV-1 protease inhibitors, GRL-142 and its analogs, markedly adapt to the structural plasticity of HIV-1 protease and exerts extreme potency with high genetic barrier
To Be Published
|
|
8W78
 
 | | Structure of Drosophila melanogaster L-2-hydroxyglutarate dehydrogenase in complex with FAD and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, DODECYL-BETA-D-MALTOSIDE, FI05204p, ... | | Authors: | Yang, J, Chen, X, Jin, S, Ding, J. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure and biochemical characterization of l-2-hydroxyglutarate dehydrogenase and its role in the pathogenesis of l-2-hydroxyglutaric aciduria.
J.Biol.Chem., 300, 2023
|
|
8W75
 
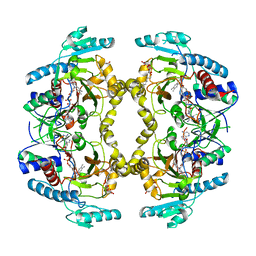 | | Structure of Drosophila melanogaster L-2-hydroxyglutarate dehydrogenase | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FI05204p, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Yang, J, Chen, X, Jin, S, Ding, J. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and biochemical characterization of l-2-hydroxyglutarate dehydrogenase and its role in the pathogenesis of l-2-hydroxyglutaric aciduria.
J.Biol.Chem., 300, 2023
|
|
8W7F
 
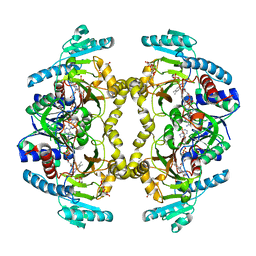 | | Structure of Drosophila melanogaster L-2-hydroxyglutarate dehydrogenase bound with FAD and a sulfate ion | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FI05204p, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yang, J, Chen, X, Jin, S, Ding, J. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structure and biochemical characterization of l-2-hydroxyglutarate dehydrogenase and its role in the pathogenesis of l-2-hydroxyglutaric aciduria.
J.Biol.Chem., 300, 2023
|
|
6LOO
 
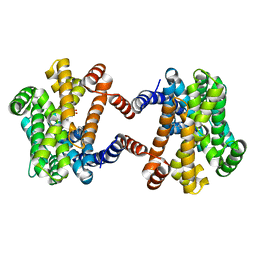 | | Crystal Structure of Class IB terpene synthase bound with geranylcitronellyl diphosphate | | Descriptor: | Tetraprenyl-beta-curcumene synthase, phosphono [(3~{R},6~{E},10~{E})-3,7,11,15-tetramethylhexadeca-6,10,14-trienyl] hydrogen phosphate, phosphono [(3~{S},6~{E},10~{E})-3,7,11,15-tetramethylhexadeca-6,10,14-trienyl] hydrogen phosphate | | Authors: | Fujihashi, M, Inagi, H, Miki, K. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Characterization of Class IB Terpene Synthase: The First Crystal Structure Bound with a Substrate Surrogate.
Acs Chem.Biol., 15, 2020
|
|
4W1Y
 
 | |
5LB2
 
 | | Apo-structure of humanised RadA-mutant humRadA2 | | Descriptor: | DNA repair and recombination protein RadA | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-15 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
4V7E
 
 | | Model of the small subunit RNA based on a 5.5 A cryo-EM map of Triticum aestivum translating 80S ribosome | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S10E, ... | | Authors: | Barrio-Garcia, C, Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | Deposit date: | 2013-11-22 | | Release date: | 2014-07-09 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion.
Nature, 506, 2014
|
|
1FCG
 
 | | ECTODOMAIN OF HUMAN FC GAMMA RECEPTOR, FCGRIIA | | Descriptor: | PROTEIN (FC RECEPTOR FC(GAMMA)RIIA) | | Authors: | Maxwell, K.F, Powell, M.S, Garrett, T.P, Hogarth, P.M. | | Deposit date: | 1999-04-07 | | Release date: | 2000-04-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the human leukocyte Fc receptor, Fc gammaRIIa.
Nat.Struct.Biol., 6, 1999
|
|
5MTS
 
 | | Complex of FimH lectin with a TazMan (thiazolylaminomannosides) family member known as potent anti-adhesive agent at 2.6 A resolution | | Descriptor: | NICKEL (II) ION, Protein FimH, [2-[[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]amino]-1,3-thiazol-5-yl]-(4-methyl-2-pyrazin-2-yl-1,3-thiazol-5-yl)methanone | | Authors: | de Ruyck, J, Bouckaert, J. | | Deposit date: | 2017-01-10 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Physiochemical Tuning of Potent Escherichia coli Anti-Adhesives by Microencapsulation and Methylene Homologation.
ChemMedChem, 12, 2017
|
|
8ADK
 
 | |
8ADJ
 
 | | Poly(ADP-ribose) glycohydrolase (PARG) from Drosophila melanogaster in complex with PARG inhibitor PDD00017272 | | Descriptor: | 1-[(2,5-dimethylpyrazol-3-yl)methyl]-N-(1-methylcyclopropyl)-3-[(2-methyl-1,3-thiazol-5-yl)methyl]-2,4-bis(oxidanylidene)quinazoline-6-sulfonamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ariza, A, Fontana, P. | | Deposit date: | 2022-07-08 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.508 Å) | | Cite: | Serine ADP-ribosylation in Drosophila provides insights into the evolution of reversible ADP-ribosylation signalling.
Nat Commun, 14, 2023
|
|
8AU4
 
 | | Structural insights reveal a heterotetramer between oncogenic K-Ras4BG12V and Rgl2, a RalA/B activator | | Descriptor: | Ral guanine nucleotide dissociation stimulator-like 2 | | Authors: | Tariq, M, Ikeya, T, Togashi, N, Fairall, L, Alejo, C.B, Kamei, S, Alonso, B.R, Campillo, M.A.M, Hudson, A, Ito, Y, Schwabe, J, Dominguez, C, Tanaka, K. | | Deposit date: | 2022-08-25 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-25 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the complex of oncogenic KRas4B G12V and Rgl2, a RalA/B activator.
Life Sci Alliance, 7, 2024
|
|
4NEY
 
 | | Crystal Structure of an engineered protein with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR277 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Engineered protein OR277, tetrabutylphosphonium | | Authors: | Guan, R, Lin, Y.-R, Koga, N, Koga, R, Castellanos, J, Seetharaman, J, Maglaqui, M, Sahdev, S, Mao, L, Xiao, R, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-10-30 | | Release date: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (2.323 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR277
To be published
|
|
1B7N
 
 | | VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Ota, M, Ogasahara, K, Yamagata, Y, Nishikawa, K, Yutani, K. | | Deposit date: | 1999-01-24 | | Release date: | 1999-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental verification of the 'stability profile of mutant protein' (SPMP) data using mutant human lysozymes.
Protein Eng., 12, 1999
|
|
4NEZ
 
 | | Crystal Structure of an engineered protein with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR276 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Engineered protein OR276, tetrabutylphosphonium | | Authors: | Guan, R, Lin, Y.-R, Koga, N, Koga, R, Castellanos, J, Seetharaman, J, Maglaqui, M, Sahdev, S, Mao, L, Xiao, R, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-10-30 | | Release date: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR276
To be published
|
|
1B7L
 
 | | VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Ota, M, Ogasahara, K, Yamagata, Y, Nishikawa, K, Yutani, K. | | Deposit date: | 1999-01-24 | | Release date: | 1999-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental verification of the 'stability profile of mutant protein' (SPMP) data using mutant human lysozymes.
Protein Eng., 12, 1999
|
|
1B7Q
 
 | | VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Ota, M, Ogasahara, K, Yamagata, Y, Nishikawa, K, Yutani, K. | | Deposit date: | 1999-01-25 | | Release date: | 1999-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Experimental verification of the 'stability profile of mutant protein' (SPMP) data using mutant human lysozymes.
Protein Eng., 12, 1999
|
|
4RJV
 
 | | Crystal Structure of a De Novo Designed Ferredoxin Fold, Northeast Structural Genomics Consortium (NESG) Target OR461 | | Descriptor: | OR461 | | Authors: | O'Connell, P.T, Lin, Y.-R, Guan, R, Koga, N, Koga, R, Seetharaman, J, Janjua, H, Xiao, R, Maglaqui, M, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-10-09 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR461
To be published
|
|
6S2N
 
 | |
6S2O
 
 | |
1B7R
 
 | | VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Ota, M, Ogasahara, K, Yamagata, Y, Nishikawa, K, Yutani, K. | | Deposit date: | 1999-01-25 | | Release date: | 1999-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental verification of the 'stability profile of mutant protein' (SPMP) data using mutant human lysozymes.
Protein Eng., 12, 1999
|
|
