8RBD
 
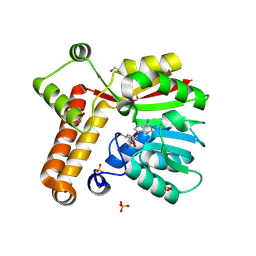 | | Crystal structure of Mycobacterium tuberculosis MmaA1 with Sinefungin (SFG) | | Descriptor: | 1,2-ETHANEDIOL, Mycolic acid methyltransferase MmaA1, SINEFUNGIN, ... | | Authors: | Kobakhidze, G, Wachelder, L, Chaudhary, B, Mazumdar, P.A, Madhurantakam, C, Dong, G. | | Deposit date: | 2023-12-04 | | Release date: | 2024-12-11 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of the mycolic acid methyl transferase 1 (MmaA1) from Mycobacterium tuberculosis in the apo-form and in complex with different cofactors reveal unique features for substrate binding.
J.Biomol.Struct.Dyn., 2025
|
|
5YHT
 
 | |
8RAQ
 
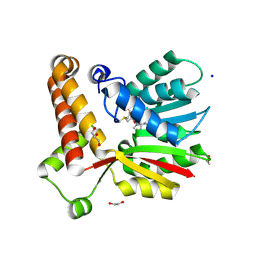 | | Crystal structure of Mycobacterium tuberculosis MmaA1 with S-adenosyl methionine (SAM) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Mycolic acid methyltransferase MmaA1, ... | | Authors: | Chaudhary, B, Kobakhidze, G, Wachelder, L, Mazumdar, P.A, Madhurantakam, C, Dong, G. | | Deposit date: | 2023-12-01 | | Release date: | 2024-12-11 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of the mycolic acid methyl transferase 1 (MmaA1) from Mycobacterium tuberculosis in the apo-form and in complex with different cofactors reveal unique features for substrate binding.
J.Biomol.Struct.Dyn., 2025
|
|
5ZON
 
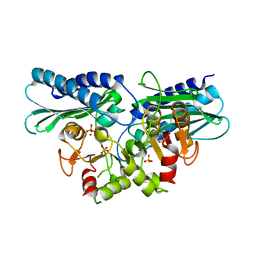 | | Histidinol phosphate phosphatase from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, Histidinol-phosphatase, PHOSPHATE ION, ... | | Authors: | Jha, B, Kumar, D, Biswal, B.K. | | Deposit date: | 2018-04-13 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Identification and structural characterization of a histidinol phosphate phosphatase from Mycobacterium tuberculosis
J. Biol. Chem., 293, 2018
|
|
5UHU
 
 | |
9FW3
 
 | |
2Y3R
 
 | | Structure of the tirandamycin-bound FAD-dependent tirandamycin oxidase TamL in P21 space group | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Carlson, J.C, Li, S, Gunatilleke, S.S, Anzai, Y, Burr, D.A, Podust, L.M, Sherman, D.H. | | Deposit date: | 2010-12-22 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Tirandamycin Biosynthesis is Mediated by Co-Dependent Oxidative Enzymes
Nat.Chem, 3, 2011
|
|
2Y3S
 
 | | Structure of the tirandamycine-bound FAD-dependent tirandamycin oxidase TamL in C2 space group | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Carlson, J.C, Li, S, Gunatilleke, S.S, Anzai, Y, Burr, D.A, Podust, L.M, Sherman, D.H. | | Deposit date: | 2010-12-23 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Tirandamycin Biosynthesis is Mediated by Co-Dependent Oxidative Enzymes
Nat.Chem, 3, 2011
|
|
2Y4G
 
 | | Structure of the Tirandamycin-bound FAD-dependent tirandamycin oxidase TamL in P212121 space group | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Carlson, J.C, Li, S, Gunatilleke, S.S, Anzai, Y, Burr, D.A, Podust, L.M, Sherman, D.H. | | Deposit date: | 2011-01-05 | | Release date: | 2011-06-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Tirandamycin Biosynthesis is Mediated by Co-Dependent Oxidative Enzymes
Nat.Chem, 3, 2011
|
|
5I4Z
 
 | | Structure of apo OmoMYC | | Descriptor: | CHLORIDE ION, GLYCEROL, Myc proto-oncogene protein, ... | | Authors: | Koelmel, W, Jung, L.A, Kuper, J, Eilers, M, Kisker, C. | | Deposit date: | 2016-02-13 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | OmoMYC blunts promoter invasion by oncogenic MYC to inhibit gene expression characteristic of MYC-dependent tumors.
Oncogene, 36, 2017
|
|
5I50
 
 | | Structure of OmoMYC bound to double-stranded DNA | | Descriptor: | DNA (5'-D(P*CP*AP*CP*CP*CP*GP*GP*TP*CP*AP*CP*GP*TP*GP*GP*CP*CP*TP*AP*CP*AP*C)-3'), DNA (5'-D(P*GP*TP*GP*TP*AP*GP*GP*CP*CP*AP*CP*GP*TP*GP*AP*CP*CP*GP*GP*GP*TP*G)-3'), Myc proto-oncogene protein | | Authors: | Koelmel, W, Jung, L.A, Kuper, J, Eilers, M, Kisker, C. | | Deposit date: | 2016-02-13 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | OmoMYC blunts promoter invasion by oncogenic MYC to inhibit gene expression characteristic of MYC-dependent tumors.
Oncogene, 36, 2017
|
|
8HCR
 
 | |
8FX9
 
 | |
6P9K
 
 | |
6FOC
 
 | | F1-ATPase from Mycobacterium smegmatis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | Zhang, T, Montgomery, M.G, Leslie, A.G.W, Cook, G.M, Walker, J.E. | | Deposit date: | 2018-02-06 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The structure of the catalytic domain of the ATP synthase fromMycobacterium smegmatisis a target for developing antitubercular drugs.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7AGR
 
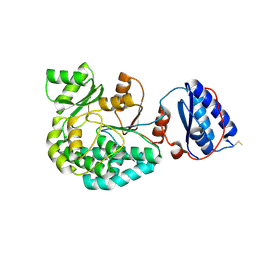 | |
7AGS
 
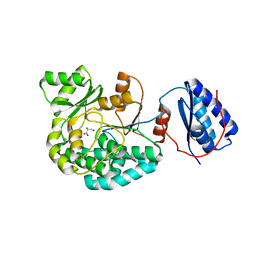 | |
7AGP
 
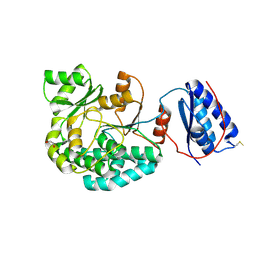 | | Structure of the AcylTransferase domain of Mycocerosic Acid Synthase from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Mycocerosic acid synthase | | Authors: | Brison, Y, Nahoum, V, Mourey, L, Maveyraud, L. | | Deposit date: | 2020-09-23 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis for Extender Unit Specificity of Mycobacterial Polyketide Synthases.
Acs Chem.Biol., 15, 2020
|
|
7AGQ
 
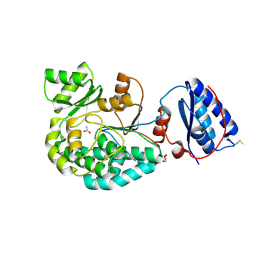 | |
7AGT
 
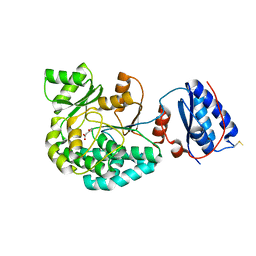 | |
7AGU
 
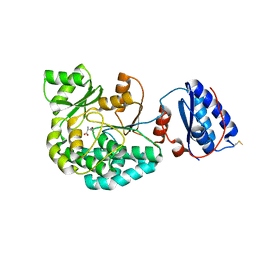 | | Structure of the S726F mutant of AcylTransferase domain of Mycocerosic Acid Synthase from Mycobacterium tuberculosis acylated with MethylMalonyl-coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, METHYLMALONIC ACID, Mycocerosic acid synthase | | Authors: | Brison, Y, Mourey, L, Maveyraud, L. | | Deposit date: | 2020-09-23 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Basis for Extender Unit Specificity of Mycobacterial Polyketide Synthases.
Acs Chem.Biol., 15, 2020
|
|
6O6P
 
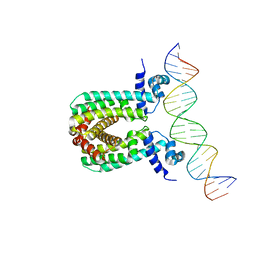 | | Structure of the regulator FasR from Mycobacterium tuberculosis in complex with DNA | | Descriptor: | DNA-forward, DNA-reverse, TetR family transcriptional regulator | | Authors: | Larrieux, N, Trajtenberg, F, Lara, J, Gramajo, H, Buschiazzo, A. | | Deposit date: | 2019-03-07 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.851 Å) | | Cite: | Mycobacterium tuberculosis FasR senses long fatty acyl-CoA through a tunnel and a hydrophobic transmission spine.
Nat Commun, 11, 2020
|
|
6O6O
 
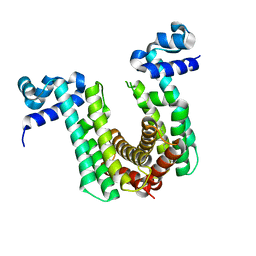 | | Structure of the regulator FasR from Mycobacterium tuberculosis | | Descriptor: | MYRISTIC ACID, TetR family transcriptional regulator | | Authors: | Larrieux, N, Trajtenberg, F, Lara, J, Gramajo, H, Buschiazzo, A. | | Deposit date: | 2019-03-07 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Mycobacterium tuberculosis FasR senses long fatty acyl-CoA through a tunnel and a hydrophobic transmission spine.
Nat Commun, 11, 2020
|
|
6O6N
 
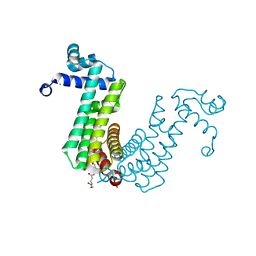 | | Structure of the regulator FasR from Mycobacterium tuberculosis in complex with C20-CoA | | Descriptor: | Arachinoyl-CoA, CHLORIDE ION, TetR family transcriptional regulator | | Authors: | Larrieux, N, Trajtenberg, F, Lara, J, Gramajo, H, Buschiazzo, A. | | Deposit date: | 2019-03-07 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mycobacterium tuberculosis FasR senses long fatty acyl-CoA through a tunnel and a hydrophobic transmission spine.
Nat Commun, 11, 2020
|
|
6GEO
 
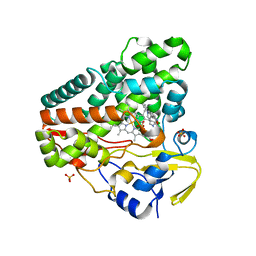 | |
